2DFY
 
 | | Crystal structure of a cyclized protein fusion of LMO4 LIM domains 1 and 2 with the LIM interacting domain of LDB1 | | Descriptor: | GLYCEROL, ZINC ION, fusion protein of LIM domain transcription factor LMO4 and LIM domain-binding protein 1 | | Authors: | Jeffries, C.M.J, Graham, S.C, Collyer, C.A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of a binary protein complex by intein-mediated cyclization
Protein Sci., 15, 2006
|
|
1U85
 
 | |
1U86
 
 | |
6PTL
 
 | |
3MMK
 
 | | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification | | Descriptor: | CHLORIDE ION, Fusion of LIM/homeobox protein Lhx4, linker, ... | | Authors: | Gadd, M.S, Langley, D.B, Guss, J.M, Matthews, J.M. | | Deposit date: | 2010-04-20 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | The structural basis for partial redundancy in a class of transcription factors, the lim-homeodomain proteins, in neural cell type specification.
J.Biol.Chem., 2011
|
|
4MJ3
 
 | | Haloalkane dehalogenase DmrA from Mycobacterium rhodesiae JS60 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, POTASSIUM ION | | Authors: | Fung, H, Gadd, M.S, Guss, J.M, Matthews, J.M. | | Deposit date: | 2013-09-03 | | Release date: | 2015-02-25 | | Last modified: | 2015-08-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and biophysical characterisation of haloalkane dehalogenases DmrA and DmrB in Mycobacterium strain JS60 and their role in growth on haloalkanes.
Mol.Microbiol., 97, 2015
|
|
3VEK
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
6CME
 
 | | Structure of wild-type ISL2-LID in complex with LHX4-LIM1+2 | | Descriptor: | LIM/homeobox protein Lhx4,Insulin gene enhancer protein ISL-2, ZINC ION | | Authors: | Stokes, P.H, Silva, A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mutation in a flexible linker modulates binding affinity for modular complexes.
Proteins, 87, 2019
|
|
3VD6
 
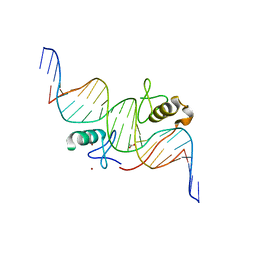 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P21 Crystal Form | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
4JCJ
 
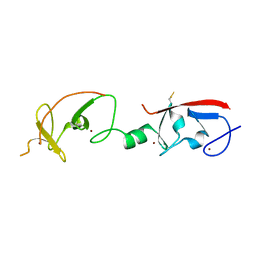 | | Crystal structure of Isl1 LIM domains with Ldb1 LIM-interaction domain | | Descriptor: | Insulin gene enhancer protein ISL-1,LIM domain-binding protein 1, ZINC ION | | Authors: | Gadd, M.S, Jacques, D.A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2013-02-21 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural basis for the regulation of the LIM-homeodomain protein islet 1 (Isl1) by intra- and intermolecular interactions.
J.Biol.Chem., 288, 2013
|
|
2RGT
 
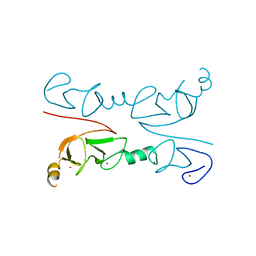 | | Crystal Structure of Lhx3 LIM domains 1 and 2 with the binding domain of Isl1 | | Descriptor: | Fusion of LIM/homeobox protein Lhx3, linker, Insulin gene enhancer protein ISL-1, ... | | Authors: | Bhati, M, Lee, M, Guss, J.M, Matthews, J.M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes.
Embo J., 27, 2008
|
|
1RUT
 
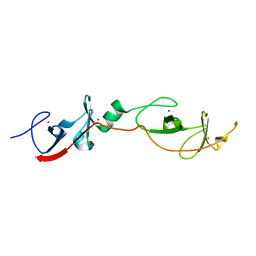 | | Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain | | Descriptor: | Fusion protein of Lmo4 protein and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Ryan, D.P, Maher, M.J, Kwan, A.H.Y, Bacca, M, Mackay, J.P, Guss, J.M, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-12-11 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex
Embo J., 23, 2004
|
|
2LXD
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for LMO2(LIM2)-Ldb1(LID) | | Descriptor: | Rhombotin-2,LIM domain-binding protein 1, ZINC ION | | Authors: | Dastmalchi, S, Wilkinson-White, L, Kwan, A.H, Gamsjaeger, R, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a tethered Lmo2(LIM2) /Ldb1(LID) complex.
Protein Sci., 21, 2012
|
|
1WO3
 
 | | Solution structure of Minimal Mutant 1 (MM1): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO6
 
 | | Solution structure of Designed Functional Finger 5 (DFF5): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO4
 
 | | Solution structure of Minimal Mutant 2 (MM2): Multiple alanine mutant of non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO5
 
 | | Solution structure of Designed Functional Finger 2 (DFF2): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1WO7
 
 | | Solution structure of Designed Functional Finger 7 (DFF7): Designed mutant based on non-native CHANCE domain | | Descriptor: | CREB Binding Protein, ZINC ION | | Authors: | Sharpe, B.K, Liew, C.K, Wilce, J.A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2004-08-12 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assessment of the robustness of a serendipitous zinc binding fold: mutagenesis and protein grafting
Structure, 13, 2005
|
|
1Y0J
 
 | | Zinc fingers as protein recognition motifs: structural basis for the GATA-1/Friend of GATA interaction | | Descriptor: | Erythroid transcription factor, ZINC ION, Zinc-finger protein ush | | Authors: | Liew, C.K, Simpson, R.J.Y, Kwan, A.H.Y, Crofts, L.A, Loughlin, F.E, Matthews, J.M, Crossley, M, Mackay, J.P. | | Deposit date: | 2004-11-15 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2JTN
 
 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
1M3V
 
 | | FLIN4: Fusion of the LIM binding domain of Ldb1 and the N-terminal LIM domain of LMO4 | | Descriptor: | ZINC ION, fusion of the LIM interacting domain of ldb1 and the N-terminal LIM domain of LMO4 | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H.Y, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2002-06-30 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
2MF8
 
 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
2JYD
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 | | Descriptor: | F5 domain of Myelin transcription factor 1, ZINC ION | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-12-12 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and biophysical analysis of the DNA binding properties of myelin transcription factor 1.
J.Biol.Chem., 283, 2008
|
|
2JX1
 
 | | Structure of the fifth zinc finger of Myelin Transcription Factor 1 in complex with RARE DNA | | Descriptor: | DNA (5'-D(*DAP*DCP*DCP*DGP*DAP*DAP*DAP*DGP*DTP*DTP*DCP*DAP*DC)-3'), DNA (5'-D(*DGP*DTP*DGP*DAP*DAP*DCP*DTP*DTP*DTP*DCP*DGP*DGP*DT)-3'), Myelin transcription factor 1 | | Authors: | Gamsjaeger, R, Swanton, M.K, Kobus, F.J, Lehtomaki, E, Lowry, J.A, Kwan, A.H, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2007-11-01 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the fifth zinc finger of Myelin Transcription Factor 1 in complex with RARE DNA
To be Published
|
|
2L4Z
 
 | | NMR structure of fusion of CtIP (641-685) to LMO4-LIM1 (18-82) | | Descriptor: | DNA endonuclease RBBP8,LIM domain transcription factor LMO4, ZINC ION | | Authors: | Liew, C, Stokes, P.H, Kwan, A.H, Matthews, J.M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction of the Breast Cancer Oncogene LMO4 with the Tumour Suppressor CtIP/RBBP8.
J.Mol.Biol., 425, 2013
|
|
