4TLN
 
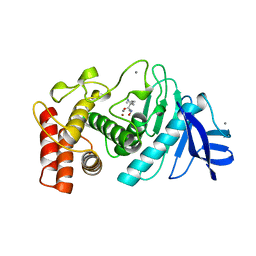 | |
7TLN
 
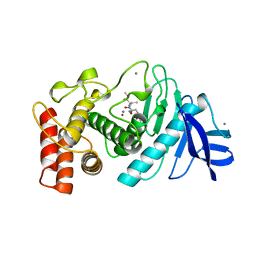 | | STRUCTURAL ANALYSIS OF THE INHIBITION OF THERMOLYSIN BY AN ACTIVE-SITE-DIRECTED IRREVERSIBLE INHIBITOR | | Descriptor: | 2-(ACETYL-HYDROXY-AMINO)-4-METHYL-PENTANOIC ACID METHYL ESTER, CALCIUM ION, THERMOLYSIN, ... | | Authors: | Matthews, B.W, Holmes, M.A, Tronrud, D.E. | | Deposit date: | 1983-01-27 | | Release date: | 1983-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the inhibition of thermolysin by an active-site-directed irreversible inhibitor.
Biochemistry, 22, 1983
|
|
5TLN
 
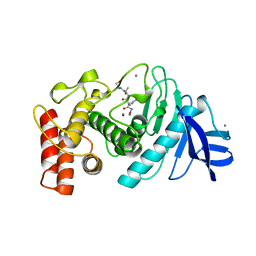 | |
1L24
 
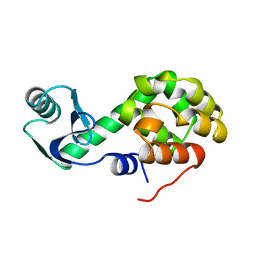 | |
1L23
 
 | |
7LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
4TMN
 
 | | SLOW-AND FAST-BINDING INHIBITORS OF THERMOLYSIN DISPLAY DIFFERENT MODES OF BINDING. CRYSTALLOGRAPHIC ANALYSIS OF EXTENDED PHOSPHONAMIDATE TRANSITION-STATE ANALOGUES | | Descriptor: | CALCIUM ION, N-[(S)-[(1R)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl](hydroxy)phosphoryl]-L-leucyl-L-alanine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
3LZM
 
 | |
5LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
6TMN
 
 | | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond | | Descriptor: | CALCIUM ION, N-[(2R,4S)-4-hydroxy-2-(2-methylpropyl)-4-oxido-7-oxo-9-phenyl-3,8-dioxa-6-aza-4-phosphanonan-1-oyl]-L-leucine, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Holden, H.M, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond.
Science, 235, 1987
|
|
8TLN
 
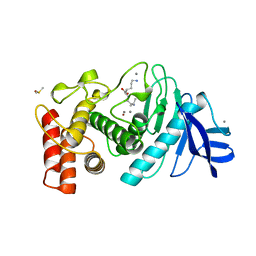 | |
4V44
 
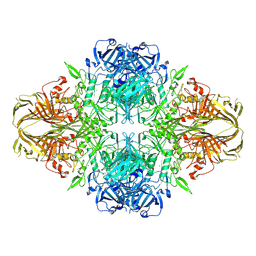 | | E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH 2-F-LACTOSE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
4V41
 
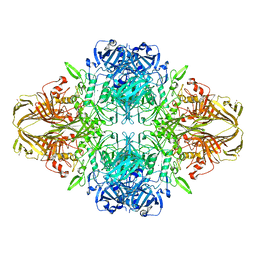 | | E. COLI (LAC Z) BETA-GALACTOSIDASE (NCS CONSTRAINED MONOMER-MONOCLINIC) | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 2000-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
4LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
3TMN
 
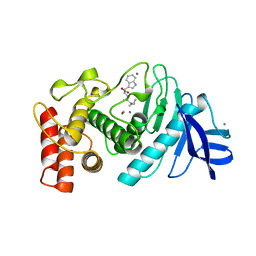 | |
4V45
 
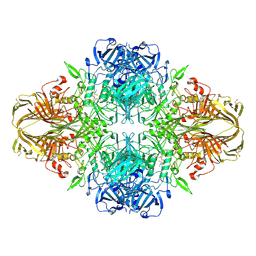 | | E. COLI (lacZ) BETA-GALACTOSIDASE-TRAPPED 2-F-GALACTOSYL-ENZYME INTERMEDIATE | | Descriptor: | 2-deoxy-2-fluoro-beta-D-galactopyranose, Beta-Galactosidase, MAGNESIUM ION, ... | | Authors: | Juers, D.H, McCarter, J.D, Withers, S.G, Matthews, B.W. | | Deposit date: | 2001-09-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Structural View of the Action of Escherichia Coli (Lacz) Beta-Galactosidase
Biochemistry, 40, 2001
|
|
5TMN
 
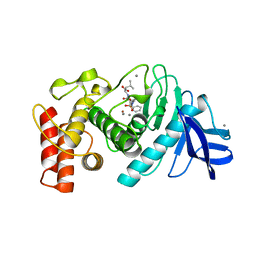 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
6CRO
 
 | |
5CRO
 
 | |
4BCL
 
 | | FMO protein from Prosthecochloris aestuarii 2K at Room Temperature | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOCHLOROPHYLL A PROTEIN | | Authors: | Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1998-04-17 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refinement of the Structure of a Water-Soluble Antenna Complex from Green Photosynthetic Bacteria by Incorporation of the Chemically Determined Amino Acid Sequence
Photosynthetic Reaction Center, 1, 1993
|
|
2TMN
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N~2~-phosphono-L-leucinamide, Thermolysin, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
3MAT
 
 | | E.COLI METHIONINE AMINOPEPTIDASE TRANSITION-STATE INHIBITOR COMPLEX | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, SODIUM ION, ... | | Authors: | Lowther, W.T, Orville, A.M, Madden, D.T, Lim, S, Rich, D.H, Matthews, B.W. | | Deposit date: | 1999-03-29 | | Release date: | 1999-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis.
Biochemistry, 38, 1999
|
|
3ORC
 
 | |
4V40
 
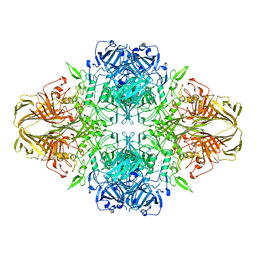 | | BETA-GALACTOSIDASE | | Descriptor: | BETA-GALACTOSIDASE, MAGNESIUM ION | | Authors: | Jacobson, R.H, Zhang, X, Dubose, R.F, Matthews, B.W. | | Deposit date: | 1994-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of beta-galactosidase from E. coli.
Nature, 369, 1994
|
|
