7T4O
 
 | |
7T4P
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO treated with potassium cyanide and copper in a native lipid nanodisc at 3.62 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4K
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.34 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4J
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.16 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4H
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.14 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4M
 
 | |
7S4L
 
 | | CryoEM structure of Methylotuvimicrobium alcaliphilum 20Z pMMO in a POPC nanodisc at 2.46 Angstrom resolution | | Descriptor: | (S)-2,3-bis(hexanoyloxy)propyl(2-(trimethylammonio)ethyl)phosphate, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, COPPER (II) ION, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
7S4I
 
 | | CryoEM structure of Methylococcus capsulatus (Bath) pMMO in a native lipid nanodisc at 2.26 Angstrom resolution | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Koo, C.W, Rosenzweig, A.C. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer.
Science, 375, 2022
|
|
5DB5
 
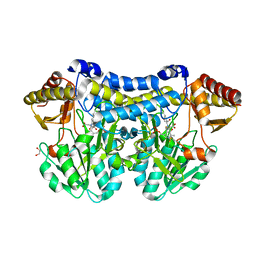 | | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, CYSTEINE, ... | | Authors: | Arbing, M.A, Shin, A, Koo, C.W, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of PLP-bound E. coli SufS (cysteine persulfide intermediate) in space group P21
To Be Published
|
|
5DBN
 
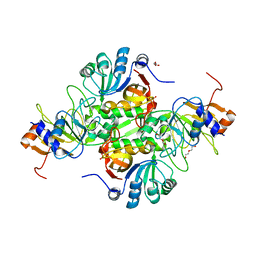 | | Crystal structure of AtoDA complex | | Descriptor: | Acetate CoA-transferase subunit alpha, Acetate CoA-transferase subunit beta, CHLORIDE ION, ... | | Authors: | Arbing, M.A, Koo, C.W, Shin, A, Medrano-Soto, A, Eisenberg, D. | | Deposit date: | 2015-08-21 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Crystal structure of AtoDA complex
To Be Published
|
|
1MDR
 
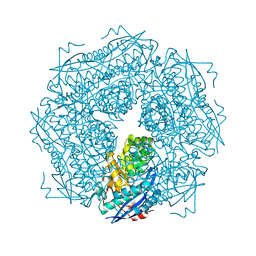 | | THE ROLE OF LYSINE 166 IN THE MECHANISM OF MANDELATE RACEMASE FROM PSEUDOMONAS PUTIDA: MECHANISTIC AND CRYSTALLOGRAPHIC EVIDENCE FOR STEREOSPECIFIC ALKYLATION BY (R)-ALPHA-PHENYLGLYCIDATE | | Descriptor: | ATROLACTIC ACID (2-PHENYL-LACTIC ACID), MAGNESIUM ION, MANDELATE RACEMASE | | Authors: | Landro, J.A, Gerlt, J.A, Kozarich, J.W, Koo, C.W, Shah, V.J, Kenyon, G.L, Neidhart, D.J, Fujita, S, Petsko, G.A. | | Deposit date: | 1993-11-19 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of lysine 166 in the mechanism of mandelate racemase from Pseudomonas putida: mechanistic and crystallographic evidence for stereospecific alkylation by (R)-alpha-phenylglycidate.
Biochemistry, 33, 1994
|
|
4RKK
 
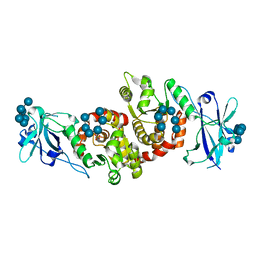 | | Structure of a product bound phosphatase | | Descriptor: | Laforin, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease.
Mol.Cell, 57, 2015
|
|
2BAY
 
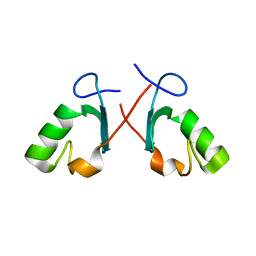 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
3BXK
 
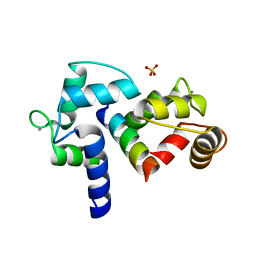 | | Crystal structure of the P/Q-type calcium channel (CaV2.1) IQ domain and CA2+calmodulin complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Mori, M.X, Vander Kooi, C.W, Leahy, D.J, Yue, D.T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the P/Q-type calcium channel (CaV2.1) IQ domain and CA2+calmodulin complex
To be Published
|
|
3BXL
 
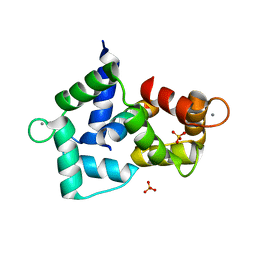 | | Crystal structure of the R-type calcium channeL (CaV2.3) IQ domain and CA2+calmodulin complex | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Mori, M.X, Vander Kooi, C.W, Leahy, D.J, Yue, D.T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin
To be Published
|
|
4KYQ
 
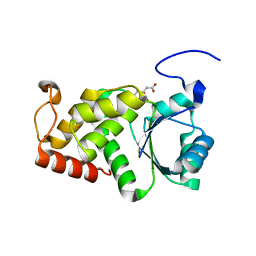 | | Structure of a product bound plant phosphatase | | Descriptor: | CITRATE ANION, Phosphoglucan phosphatase LSF2, chloroplastic | | Authors: | Meekins, D.A, Guo, H.-F, Husodo, S, Paasch, B.C, Bridges, T.M, Santelia, D, Kotting, O, Vander Kooi, C.W, Gentry, M.S. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of the Arabidopsis Glucan Phosphatase LIKE SEX FOUR2 Reveals a Unique Mechanism for Starch Dephosphorylation.
Plant Cell, 25, 2013
|
|
4KYR
 
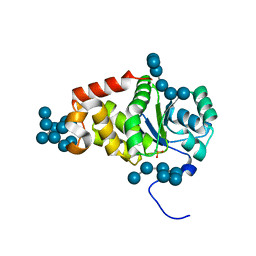 | | Structure of a product bound plant phosphatase | | Descriptor: | PHOSPHATE ION, Phosphoglucan phosphatase LSF2, chloroplastic, ... | | Authors: | Meekins, D.A, Guo, H.-F, Husodo, S, Paasch, B.C, Bridges, T.M, Santelia, D, Kotting, O, Vander Kooi, C.W, Gentry, M.S. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis Glucan Phosphatase LIKE SEX FOUR2 Reveals a Unique Mechanism for Starch Dephosphorylation.
Plant Cell, 25, 2013
|
|
3LRV
 
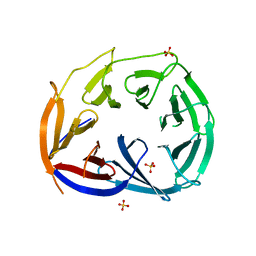 | |
3NME
 
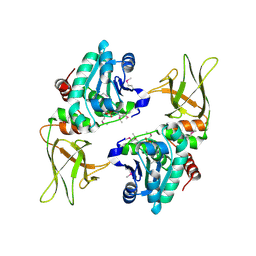 | | Structure of a plant phosphatase | | Descriptor: | PHOSPHATE ION, SEX4 glucan phosphatase | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2010-06-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the glucan phosphatase activity of Starch Excess4.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4DEQ
 
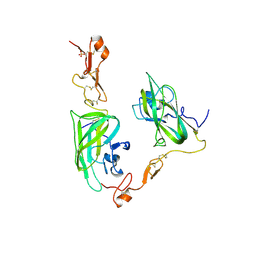 | | Structure of the Neuropilin-1/VEGF-A complex | | Descriptor: | Neuropilin-1, Vascular endothelial growth factor A, PHOSPHATE ION | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2012-01-21 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Structural Basis for Selective Vascular Endothelial Growth Factor-A (VEGF-A) Binding to Neuropilin-1.
J.Biol.Chem., 287, 2012
|
|
2ORX
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1 | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2ORZ
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1, Tuftsin | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1N87
 
 | |
5C1F
 
 | | Structure of the Imp2 F-BAR domain | | Descriptor: | FORMIC ACID, Septation protein imp2 | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2015-06-13 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3551 Å) | | Cite: | The Tubulation Activity of a Fission Yeast F-BAR Protein Is Dispensable for Its Function in Cytokinesis.
Cell Rep, 14, 2016
|
|
4QDQ
 
 | | Physical basis for Nrp2 ligand binding | | Descriptor: | GLYCEROL, Neuropilin-2, SULFATE ION | | Authors: | Parker, M.W, Vander Kooi, C.W. | | Deposit date: | 2014-05-14 | | Release date: | 2015-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for VEGF-C Binding to Neuropilin-2 and Sequestration by a Soluble Splice Form.
Structure, 23, 2015
|
|
