2RML
 
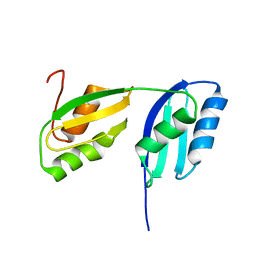 | | Solution structure of the N-terminal soluble domains of Bacillus subtilis CopA | | Descriptor: | Copper-transporting P-type ATPase copA | | Authors: | Singleton, C, Banci, L, Bertini, I, Ciofi-Baffoni, S, Tenori, L, Kihlken, M.A, Boetzel, R, Le Brun, N.E. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Cu(I)-binding properties of the N-terminal soluble domains of Bacillus subtilis CopA
Biochem.J., 411, 2008
|
|
7OZM
 
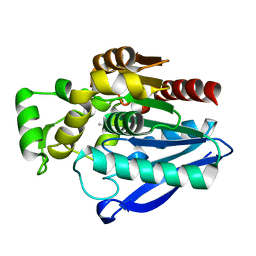 | | Crystal Structure of mtbMGL K74A (Closed Cap Conformation) | | Descriptor: | ISOPROPYL ALCOHOL, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-28 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
7P0Y
 
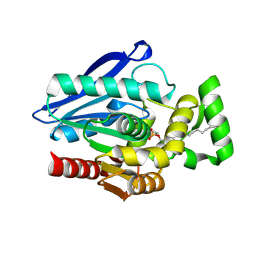 | | Crystal Structure of mtbMGL K74A (Substrate Analog Complex) | | Descriptor: | 1-[butyl(fluoranyl)phosphoryl]oxyhexadecane, Monoacylglycerol lipase | | Authors: | Grininger, C, Aschauer, P, Pavkov-Keller, T, Oberer, M. | | Deposit date: | 2021-06-30 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Changes in the Cap of Rv0183/mtbMGL Modulate the Shape of the Binding Pocket.
Biomolecules, 11, 2021
|
|
7OTT
 
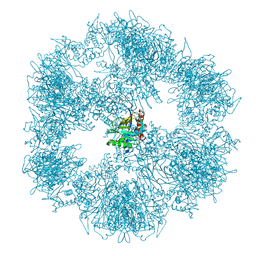 | |
3PQI
 
 | |
3PQH
 
 | |
2X5N
 
 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
7BJV
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI09181 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
7BJU
 
 | | Crystal structure of the ligand-binding domains of the heterodimer EcR/USP bound to the synthetic agonist BYI08346 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DI(HYDROXYETHYL)ETHER, Ecdysone Receptor, ... | | Authors: | Browning, C, McEwen, A.G, Billas, I.M.L. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Nonsteroidal ecdysone receptor agonists use a water channel for binding to the ecdysone receptor complex EcR/USP.
J Pestic Sci, 46, 2021
|
|
3QR8
 
 | |
1UR6
 
 | | NMR based structural model of the UbcH5B-CNOT4 complex | | Descriptor: | POTENTIAL TRANSCRIPTIONAL REPRESSOR NOT4HP, UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2, ZINC ION | | Authors: | Dominguez, C, Bonvin, A.M.J.J, Winkler, G.S, Van Schaik, F.M.A, Timmers, H.Th.M, Boelens, R. | | Deposit date: | 2003-10-27 | | Release date: | 2004-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, THEORETICAL MODEL | | Cite: | Structural Model of the Ubch5B/Cnot4 Complex Revealed by Combining NMR, Mutagenesis, and Docking Approaches.
Structure, 12, 2004
|
|
3QR7
 
 | | Crystal structure of the C-terminal fragment of the bacteriophage P2 membrane-piercing protein gpV | | Descriptor: | Baseplate assembly protein V, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Browning, C, Shneider, M, Leiman, P.G. | | Deposit date: | 2011-02-17 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Phage pierces the host cell membrane with the iron-loaded spike.
Structure, 20, 2012
|
|
4RU3
 
 | |
1RHF
 
 | | Crystal Structure of human Tyro3-D1D2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, Tyrosine-protein kinase receptor TYRO3, ... | | Authors: | Heiring, C, Dahlback, B, Muller, Y.A. | | Deposit date: | 2003-11-14 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand recognition and homophilic interactions in Tyro3: structural insights into the Axl/Tyro3 receptor tyrosine kinase family.
J.Biol.Chem., 279, 2004
|
|
2HGM
 
 | | NMR structure of the second qRRM domain of human hnRNP F | | Descriptor: | Heterogeneous nuclear ribonucleoprotein F | | Authors: | Dominguez, C, Allain, F.H.-T. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition.
Nucleic Acids Res., 34, 2006
|
|
2HGL
 
 | | NMR structure of the first qRRM domain of human hnRNP F | | Descriptor: | Heterogeneous nuclear ribonucleoprotein F | | Authors: | Dominguez, C, Allain, F.H.-T. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition.
Nucleic Acids Res., 34, 2006
|
|
2HGN
 
 | | NMR structure of the third qRRM domain of human hnRNP F | | Descriptor: | Heterogeneous nuclear ribonucleoprotein F | | Authors: | Dominguez, C, Allain, F.H.-T. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition.
Nucleic Acids Res., 34, 2006
|
|
1C16
 
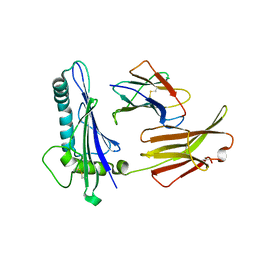 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | Descriptor: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | Authors: | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | Deposit date: | 1999-07-20 | | Release date: | 2000-01-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
4B1H
 
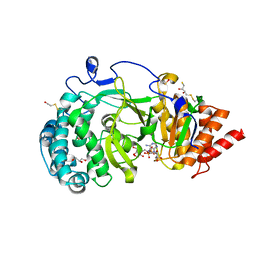 | | Structure of human PARG catalytic domain in complex with ADP-ribose | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
4B1J
 
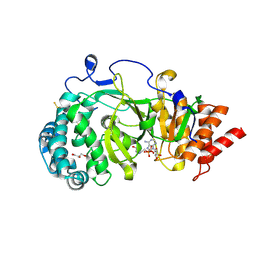 | | Structure of human PARG catalytic domain in complex with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, McAlister, M, Overman, R, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
4B1I
 
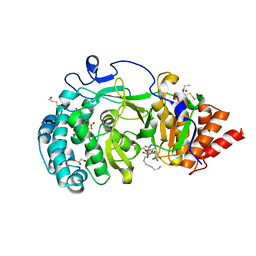 | | Structure of human PARG catalytic domain in complex with OA-ADP-HPD | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 8-n-octylamino-adenosine diphosphate hydroxypyrrolidinediol, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, Johnson, T, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
8RO4
 
 | | The crystal structure of 2-hydroxy-3-keto-glucal hydratase AtHYD from A. tumefaciens | | Descriptor: | 2-hydroxy-3-keto-glucal hydratase, MANGANESE (II) ION | | Authors: | Grininger, C, Bitter, J, Pfeiffer, M, Nidetzky, B, Pavkov-Keller, T. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 2024
|
|
6PT0
 
 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
9FS9
 
 | |
9FSA
 
 | |
