6OKB
 
 | | Prohead 2 of the phage T5 | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-12 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OMA
 
 | |
6OMC
 
 | | capsid of T5 virion | | Descriptor: | Major capsid protein | | Authors: | Huet, A, Duda, R.L, Boulanger, P, Conway, J.F. | | Deposit date: | 2019-04-18 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8FQL
 
 | | Portal vertex of HK97 phage | | Descriptor: | Portal protein | | Authors: | Huet, A, Oh, B, Maurer, J, Duda, R.L, Conway, J.F. | | Deposit date: | 2023-01-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A symmetry mismatch unraveled: How phage HK97 scaffold flexibly accommodates a 12-fold pore at a 5-fold viral capsid vertex.
Sci Adv, 9, 2023
|
|
8FQK
 
 | | Asymmetric unit of HK97 phage prohead I | | Descriptor: | Scaffolding domain delta | | Authors: | Huet, A, Oh, B, Maurer, J, Duda, R.L, Conway, J.F. | | Deposit date: | 2023-01-06 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A symmetry mismatch unraveled: How phage HK97 scaffold flexibly accommodates a 12-fold pore at a 5-fold viral capsid vertex.
Sci Adv, 9, 2023
|
|
4ULW
 
 | |
2WBU
 
 | | CRYSTAL STRUCTURE OF THE ZINC FINGER DOMAIN OF KLF4 BOUND TO ITS TARGET DNA | | Descriptor: | 5'-D(*DGP*DAP*DGP*DGP*DCP*DGP*DTP* DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DAP*DCP*DGP*DCP* DCP*DTP*DC)-3', KRUEPPEL-LIKE FACTOR 4, ... | | Authors: | Schuetz, A, Zocher, G, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-05 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
5N7H
 
 | |
5N7K
 
 | |
5N7I
 
 | |
6EKU
 
 | | Vibrio cholerae neuraminidase complexed with zanamivir | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
6EKS
 
 | | Vibrio cholerae neuraminidase complexed with oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Schuetz, A, Heinemann, U. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Different Inhibitory Potencies of Oseltamivir Carboxylate, Zanamivir, and Several Tannins on Bacterial and Viral Neuraminidases as Assessed in a Cell-Free Fluorescence-Based Enzyme Inhibition Assay.
Molecules, 22, 2017
|
|
3C0Y
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 | | Descriptor: | Histone deacetylase 7a, POTASSIUM ION, ZINC ION | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Loppnau, P, Kwiatkowski, N.P, Mazitschek, R, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
3C10
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 in complex with Trichostatin A (TSA) | | Descriptor: | Histone deacetylase 7a, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Min, J, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
3C0Z
 
 | | Crystal structure of catalytic domain of human histone deacetylase HDAC7 in complex with SAHA | | Descriptor: | Histone deacetylase 7a, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, POTASSIUM ION, ... | | Authors: | Min, J, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity.
J.Biol.Chem., 283, 2008
|
|
1Z4R
 
 | | Human GCN5 Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, General control of amino acid synthesis protein 5-like 2 | | Authors: | Dong, A, Bernstein, G, Schuetz, A, Antoshenko, T, Wu, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A
Proteins, 68, 2007
|
|
2H9M
 
 | | WDR5 in complex with unmodified H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2GNQ
 
 | | Structure of wdr5 | | Descriptor: | CHLORIDE ION, WD-repeat protein 5 | | Authors: | Min, J, Schuetz, A, Allali-Hassani, A, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-10 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H9N
 
 | | WDR5 in complex with monomethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2PSW
 
 | | Human MAK3 homolog in complex with CoA | | Descriptor: | COENZYME A, N-acetyltransferase 13 | | Authors: | Walker, J.R, Schuetz, A, Antoshenko, T, Wu, H, Bernstein, G, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Human MAK3 homolog.
To be Published
|
|
2H9L
 
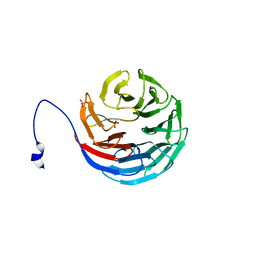 | | WDR5delta23 | | Descriptor: | SULFATE ION, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2H9P
 
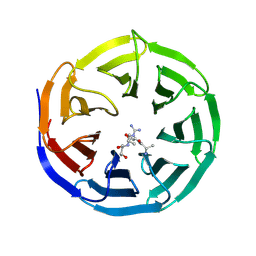 | | WDR5 in complex with trimethylated H3K4 peptide | | Descriptor: | H3 histone, WD-repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-10 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for molecular recognition and presentation of histone H3 By WDR5.
Embo J., 25, 2006
|
|
2O9K
 
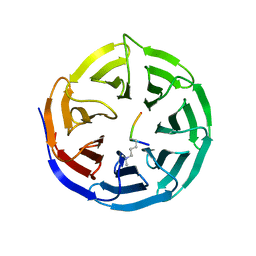 | | WDR5 in Complex with Dimethylated H3K4 Peptide | | Descriptor: | H3 HISTONE, WD repeat protein 5 | | Authors: | Min, J.R, Schuetz, A, Allali-Hassani, A, Martin, F, Loppnau, P, Vedadi, M, Weigelt, J, Sundstrom, M, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Molecular Recognition and Presentation of Histone H3 by Wdr5.
Embo J., 25, 2006
|
|
3TT9
 
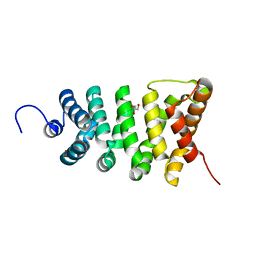 | | Crystal structure of the stable degradation fragment of human plakophilin 2 isoform a (PKP2a) C752R variant | | Descriptor: | GLYCEROL, Plakophilin-2 | | Authors: | Schuetz, A, Roske, Y, Gerull, B, Heinemann, U. | | Deposit date: | 2011-09-14 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations.
Circ Cardiovasc Genet, 5, 2012
|
|
2GL8
 
 | | Human Retinoic acid receptor RXR-gamma ligand-binding domain | | Descriptor: | Retinoic acid receptor RXR-gamma | | Authors: | Min, J.R, Schuetz, A, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-04 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Retinoic acid receptor RXR-gamma ligand-binding domain.
To be Published
|
|
