4UFT
 
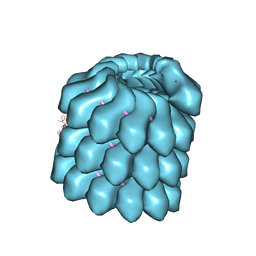 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
9E0O
 
 | | CryoEM structure of inducible Lysine decarboxylase from Hafnia alvei L-hydrazino-Lysine analog at 2.04 Angstrom resolution | | Descriptor: | (2R)-6-amino-2-[(2E)-2-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)hydrazin-1-yl]hexanoic acid, Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
9E0M
 
 | | CryoEM structure of holoenzyme of inducible Lysine decarboxylase from Hafnia alvei holoenzyme at 2.19 Angstrom resolution | | Descriptor: | Lysine decarboxylase, inducible | | Authors: | Duhoo, Y, Desfosses, A, Gutsche, I, Doukov, T.I, Berkowitz, D.B. | | Deposit date: | 2024-10-18 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | alpha-Hydrazino Acids Inhibit Pyridoxal Phosphate-Dependent Decarboxylases via "Catalytically Correct" Ketoenamine Tautomers: A Special Motif for Chemical Biology and Drug Discovery?
Acs Catalysis, 15, 2025
|
|
5FL2
 
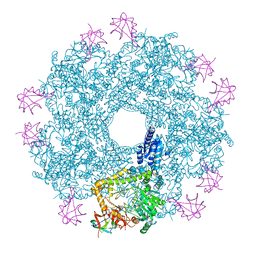 | | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
3K2S
 
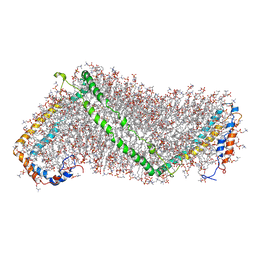 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
8PHE
 
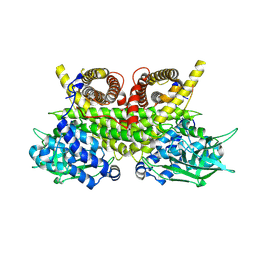 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8OP2
 
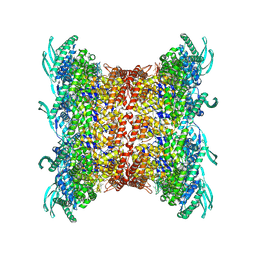 | |
8OP1
 
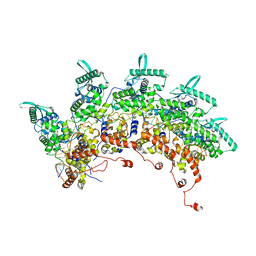 | | Subsection of a helical nucleocapsid of the Respiratory Syncytial Virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Gonnin, L, Desfosses, A, Eleouet, J.F, Galloux, M, Gutsche, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural landscape of the respiratory syncytial virus nucleocapsids.
Nat Commun, 14, 2023
|
|
8OOU
 
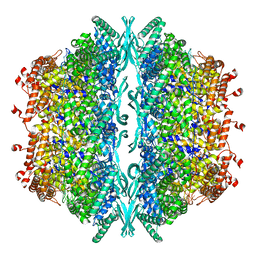 | |
4UPB
 
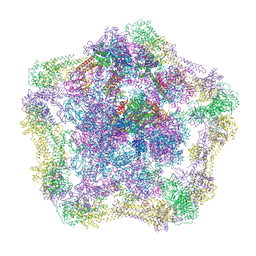 | | Electron cryo-microscopy of the complex formed between the hexameric ATPase RavA and the decameric inducible decarboxylase LdcI | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-15 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
4UPF
 
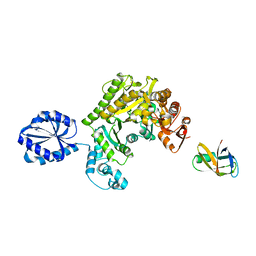 | | Assembly principles of the unique cage formed by the ATPase RavA hexamer and the lysine decarboxylase LdcI decamer | | Descriptor: | ATPASE RAVA, LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Malet, H, Liu, K, El Bakkouri, M, Chan, S.W.S, Effantin, G, Bacia, M, Houry, W.A, Gutsche, I. | | Deposit date: | 2014-06-16 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Assembly Principles of a Unique Cage Formed by Hexameric and Decameric E. Coli Proteins.
Elife, 3, 2014
|
|
4ZC0
 
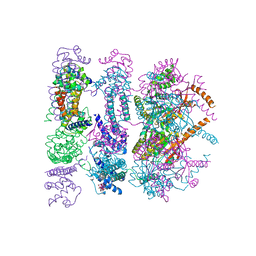 | | Structure of a dodecameric bacterial helicase | | Descriptor: | HEXATANTALUM DODECABROMIDE, Replicative DNA helicase | | Authors: | Bazin, A, Cherrier, M.V, Gutsche, I, Timmins, J, Terradot, L. | | Deposit date: | 2015-04-15 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Structure and primase-mediated activation of a bacterial dodecameric replicative helicase.
Nucleic Acids Res., 43, 2015
|
|
1WCE
 
 | | Crystal structure of the T13 IBDV viral particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
1WCD
 
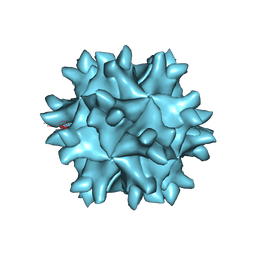 | | Crystal structure of IBDV T1 virus-like particle reveals a missing link in icosahedral viruses evolution | | Descriptor: | MAJOR STRUCTURAL PROTEIN VP2 | | Authors: | Coulibaly, F, Chevalier, C, Gutsche, I, Pous, J, Bressanelli, S, Navaza, J, Delmas, B, Rey, F.A. | | Deposit date: | 2004-11-12 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Birnavirus Crystal Structure Reveals Structural Relationships Among Icosahedral Viruses.
Cell(Cambridge,Mass.), 120, 2005
|
|
5FKX
 
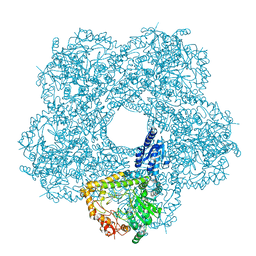 | | Structure of E.coli inducible lysine decarboxylase at active pH | | Descriptor: | LYSINE DECARBOXYLASE, INDUCIBLE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
5FKZ
 
 | | Structure of E.coli Constitutive lysine decarboxylase | | Descriptor: | LYSINE DECARBOXYLASE, CONSTITUTIVE | | Authors: | Kandiah, E, Carriel, D, Perard, J, Malet, H, Bacia, M, Liu, K, Chan, S.W.S, Houry, W.A, Ollagnier de Choudens, S, Elsen, S, Gutsche, I. | | Deposit date: | 2015-10-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Insights Into the Escherichia Coli Lysine Decarboxylases and Molecular Determinants of Interaction with the Aaa+ ATPase Rava.
Sci.Rep., 6, 2016
|
|
6Y3X
 
 | |
6YN5
 
 | | Inducible lysine decarboxylase LdcI decamer, pH 7.0 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Jessop, M, Felix, J, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6YN6
 
 | | Inducible lysine decarboxylase LdcI stacks, pH 5.7 | | Descriptor: | Inducible lysine decarboxylase | | Authors: | Felix, J, Jessop, M, Desfosses, A, Effantin, G, Gutsche, I. | | Deposit date: | 2020-04-10 | | Release date: | 2021-01-13 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Supramolecular assembly of the Escherichia coli LdcI upon acid stress.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Q7M
 
 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
6Q6I
 
 | |
6Q7L
 
 | | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RavA, Inducible lysine decarboxylase, ... | | Authors: | Arragain, B, Felix, J, Malet, H, Gutsche, I, Jessop, M. | | Deposit date: | 2018-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex.
Commun Biol, 3, 2020
|
|
2WJY
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 Orthorhombic form | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, SULFATE ION, ZINC ION | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
2WJV
 
 | | Crystal structure of the complex between human nonsense mediated decay factors UPF1 and UPF2 | | Descriptor: | REGULATOR OF NONSENSE TRANSCRIPTS 1, REGULATOR OF NONSENSE TRANSCRIPTS 2, SULFATE ION, ... | | Authors: | Clerici, M, Mourao, A, Gutsche, I, Gehring, N.H, Hentze, M.W, Kulozik, A, Kadlec, J, Sattler, M, Cusack, S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unusual Bipartite Mode of Interaction between the Nonsense-Mediated Decay Factors, Upf1 and Upf2.
Embo J., 28, 2009
|
|
