1NEV
 
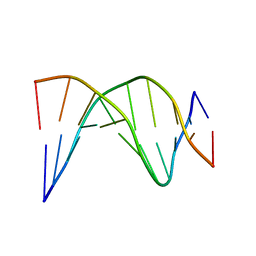 | | A-tract decamer | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*AP*AP*AP*CP*GP*G)-3' | | Authors: | Barbic, A, Zimmer, D.P, Crothers, D.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural origins of adenine-tract bending
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7OTI
 
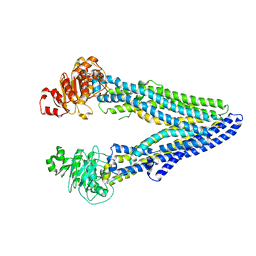 | | Structure of ABCB1/P-glycoprotein in apo state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Barbieri, A, Thonghin, N, Shafi, T, Prince, S.M, Collins, R.F. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor.
Membranes (Basel), 11, 2021
|
|
7OTG
 
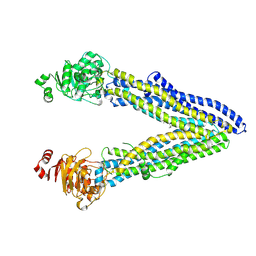 | | Structure of ABCB1/P-glycoprotein in the presence of the CFTR potentiator ivacaftor | | Descriptor: | Multidrug resistance protein 1A, N-(2,4-di-tert-butyl-5-hydroxyphenyl)-4-oxo-1,4-dihydroquinoline-3-carboxamide | | Authors: | Ford, R.C, Barbieri, A, Thonghin, N, Shafi, T, Prince, S.M, Collins, R.F. | | Deposit date: | 2021-06-10 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structure of ABCB1/P-Glycoprotein in the Presence of the CFTR Potentiator Ivacaftor.
Membranes (Basel), 11, 2021
|
|
6BIQ
 
 | |
6BIO
 
 | |
6BIM
 
 | |
2IX1
 
 | | RNase II D209N mutant | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP *AP*AP*A)-3', EXORIBONUCLEASE 2, MAGNESIUM ION | | Authors: | Frazao, C, McVey, C.E, Amblar, M, Barbas, A, Vonrhein, C, Arraiano, C.M, Carrondo, M.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Unravelling the Dynamics of RNA Degradation by Ribonuclease II and its RNA-Bound Complex
Nature, 443, 2006
|
|
2IX0
 
 | | RNase II | | Descriptor: | CALCIUM ION, CYTIDINE-5'-MONOPHOSPHATE, EXORIBONUCLEASE 2, ... | | Authors: | Frazao, C, Mcvey, C.E, Amblar, M, Barbas, A, Vonrhein, C, Arraiano, C.M, Carrondo, M.A. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Unravelling the Dynamics of RNA Degradation by Ribonuclease II and its RNA-Bound Complex
Nature, 7, 2006
|
|
6Q81
 
 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P-glycoprotein (ABCB1) | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-12-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
4UU4
 
 | | Crystal structure of LptH, the LptA homologous periplasmic component of the conserved lipopolysaccharide transport device from Pseudomonas aeruginosa | | Descriptor: | PERIPLASMIC LIPOPOLYSACCHARIDE TRANSPORT PROTEIN LPTH | | Authors: | Bollati, M, Villa, R, Gourlay, L.J, Barbiroli, A, Deho, G, Benedet, M, Polissi, A, Martorana, A, Sperandeo, P, Bolognesi, M, Nardini, M. | | Deposit date: | 2014-07-24 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Crystal Structure of Lpth, the Periplasmic Component of the Lipopolysaccharide Transport Machinery from Pseudomonas Aeruginosa.
FEBS J., 282, 2015
|
|
6Y2K
 
 | | Crystal structure of beta-galactosidase from the psychrophilic Marinomonas ef1 | | Descriptor: | CHLORIDE ION, GLYCEROL, beta-galactosidase | | Authors: | Mangiagalli, M, Lapi, M, Maione, S, Orlando, M, Brocca, S, Pesce, A, Barbiroli, A, Pucciarelli, S, Camilloni, C, Lotti, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement.
Febs J., 288, 2021
|
|
5L8S
 
 | | The crystal structure of a cold-adapted acylaminoacyl peptidase reveals a novel quaternary architecture based on the arm-exchange mechanism | | Descriptor: | Amino acyl peptidase, SULFATE ION | | Authors: | Brocca, S, Ferrari, C, Barbiroli, A, Pesce, A, Lotti, M, Nardini, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A bacterial acyl aminoacyl peptidase couples flexibility and stability as a result of cold adaptation.
FEBS J., 283, 2016
|
|
6GDI
 
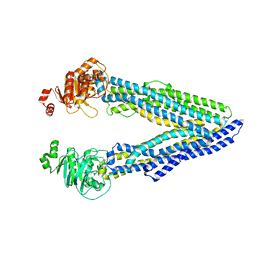 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
6BHQ
 
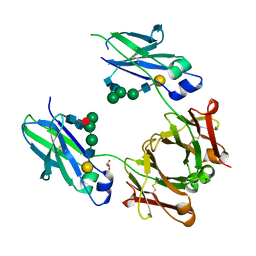 | | Mouse Immunoglobulin G 2c Fc fragment with complex-type glycan | | Descriptor: | GLYCEROL, Igh protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Falconer, D, Barb, A. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1.
PLoS ONE, 13, 2018
|
|
6BHY
 
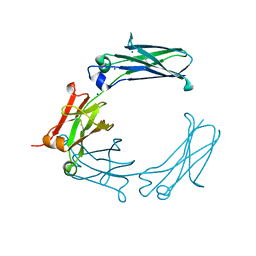 | | Mouse Immunoglobulin G 2c Fc fragment with single GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, SODIUM ION | | Authors: | Falconer, D, Barb, A. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mouse IgG2c Fc loop residues promote greater receptor-binding affinity than mouse IgG2b or human IgG1.
PLoS ONE, 13, 2018
|
|
4RMW
 
 | |
4RMU
 
 | |
4RMV
 
 | |
8EV5
 
 | | NlpC B3 covalently bound with E64 inhibitor fragment | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase, ... | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
8EV4
 
 | | NlpC B3 - Trichomonas Vaginalis | | Descriptor: | Clan CA, family C40, NlpC/P60 superfamily cysteine peptidase | | Authors: | Barnett, M.J, Goldstone, D.C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | NlpC/P60 peptidoglycan hydrolases of Trichomonas vaginalis have complementary activities that empower the protozoan to control host-protective lactobacilli.
Plos Pathog., 19, 2023
|
|
5CSG
 
 | |
5CSB
 
 | | The crystal structure of beta2-microglobulin D76N mutant at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
5CS7
 
 | | The crystal structure of wt beta2-microglobulin at room temperature | | Descriptor: | Beta-2-microglobulin | | Authors: | de Rosa, M, Mota, C.S, de Sanctis, D, Bolognesi, M, Ricagno, S. | | Deposit date: | 2015-07-23 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity.
Nat Commun, 9, 2018
|
|
6I8C
 
 | | Crystal structure of the murine beta-2-microglobulin. | | Descriptor: | Beta-2-microglobulin, TRIETHYLENE GLYCOL | | Authors: | Achour, A, Sandalova, T, Ricagno, S, Sun, R. | | Deposit date: | 2018-11-20 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and biophysical comparison of human and mouse beta-2 microglobulin reveals the molecular determinants of low amyloid propensity.
Febs J., 287, 2020
|
|
6GRZ
 
 | |
