6Y9T
 
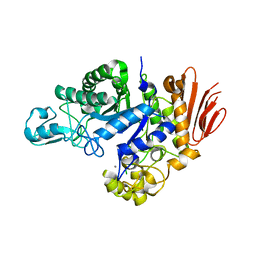 | | Family GH13_31 enzyme | | Descriptor: | Alpha-glucosidase, CALCIUM ION | | Authors: | Andersen, S, Poulsen, J.C.N, Moeller, M.S, Abou Hachem, M, Lo Leggio, L. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | An 1,4-alpha-Glucosyltransferase Defines a New Maltodextrin Catabolism Scheme in Lactobacillus acidophilus.
Appl.Environ.Microbiol., 86, 2020
|
|
1YRX
 
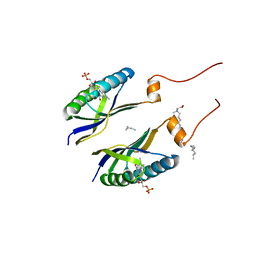 | | Structure of a novel photoreceptor: the BLUF domain of AppA from Rhodobacter sphaeroides | | Descriptor: | FLAVIN MONONUCLEOTIDE, N-DODECYL-N,N-DIMETHYLGLYCINATE, hypothetical protein Rsph03001874 | | Authors: | Anderson, S, Dragnea, V, Masuda, S, Ybe, J, Moffat, K, Bauer, C. | | Deposit date: | 2005-02-05 | | Release date: | 2005-06-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Novel Photoreceptor, the BLUF Domain of AppA from Rhodobacter sphaeroides
Biochemistry, 44, 2005
|
|
1S1Y
 
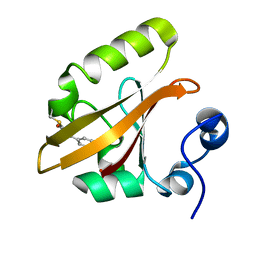 | | Photoactivated chromophore conformation in Photoactive Yellow Protein (E46Q mutant) from 10 microseconds to 3 milliseconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Srajer, V, Pahl, R, Rajagopal, S, Schotte, F, Anfinrud, P, Wulff, M, Moffat, K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein
Structure, 12, 2004
|
|
1S1Z
 
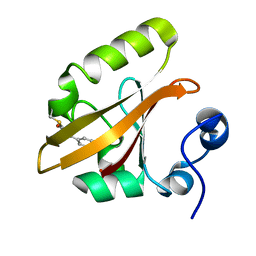 | | Photoactivated chromophore conformation in Photoactive Yellow Protein (E46Q mutant) from 10 to 500 nanoseconds | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive Yellow Protein | | Authors: | Anderson, S, Srajer, V, Pahl, R, Rajagopal, S, Schotte, F, Anfinrud, P, Wulff, M, Moffat, K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chromophore conformation and the evolution of tertiary structural changes in photoactive yellow protein
Structure, 12, 2004
|
|
1OTA
 
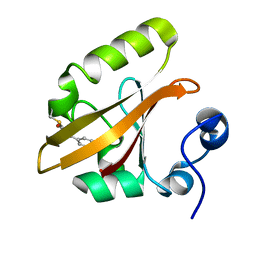 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTB
 
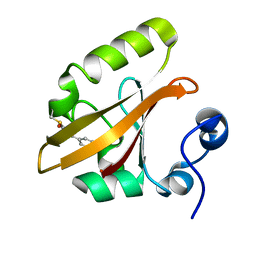 | | WILD TYPE PHOTOACTIVE YELLOW PROTEIN, P63 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OT6
 
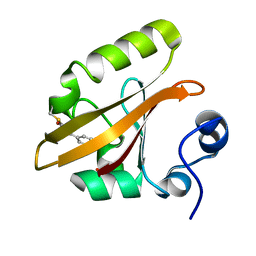 | |
1OT9
 
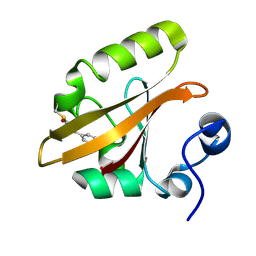 | | CRYOTRAPPED STATE IN WILD TYPE PHOTOACTIVE YELLOW PROTEIN, INDUCED WITH CONTINUOUS ILLUMINATION AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTI
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 295K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OTD
 
 | |
1OTE
 
 | | E46Q MUTANT OF PHOTOACTIVE YELLOW PROTEIN, P65 AT 110K | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, photoactive yellow protein, PYP | | Authors: | Anderson, S, Crosson, S, Moffat, K. | | Deposit date: | 2003-03-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Short hydrogen bonds in photoactive yellow protein.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6GF1
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | SULFATE ION, Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
6GF2
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
1TS7
 
 | | Structure of the pR cis wobble and pR E46Q intermediates from time-resolved Laue crystallography | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Ihee, H, Rajagopal, S, Srajer, V, Pahl, R, Anderson, S, Schmidt, M, Schotte, F, Anfinrud, P.A, Wulff, M, Moffat, K. | | Deposit date: | 2004-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Visualizing reaction pathways in photoactive yellow protein from nanoseconds to seconds.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4MLG
 
 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
1A80
 
 | | Native 2,5-DIKETO-D-GLUCONIC acid reductase a from CORYNBACTERIUM SP. complexed with nadph | | Descriptor: | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khurana, S, Powers, D.B, Anderson, S, Blaber, M. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2SGP
 
 | | PRO 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | OVOMUCOID INHIBITOR, PHOSPHATE ION, PROTEINASE B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
2SGQ
 
 | | GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pH's
To be Published
|
|
2SGF
 
 | | PHE 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B | | Descriptor: | Ovomucoid, PHOSPHATE ION, Streptogrisin B | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
2SGD
 
 | | ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 | | Descriptor: | Ovomucoid, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Huang, K, Lu, W, Anderson, S, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 1999-03-25 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Recruitment of a Buried K+ Ion to Stabilize the Negative Charge of Ionized P1 in the Hydrophobic Pocket: Crystal Structures of Glu18, Gln18, Asp18 and Asn18 Variants of Turkey Ovomucoid Inhibitor Third Domain Complexed with Streptomyces griseus Protease B at Various pHs
To be Published
|
|
2HFN
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | FLAVIN MONONUCLEOTIDE, Synechocystis Photoreceptor (Slr1694) | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
2HFO
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | Activator of photopigment and puc expression, FLAVIN MONONUCLEOTIDE | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
5EQ4
 
 | | Crystal structure of the SrpA adhesin R347E mutant from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ2
 
 | | Crystal Structure of the SrpA Adhesin from Streptococcus sanguinis | | Descriptor: | ACETATE ION, CALCIUM ION, Platelet-binding glycoprotein | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
5EQ3
 
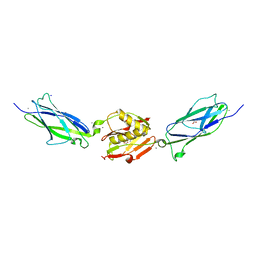 | | Crystal structure of the SrpA adhesin from Streptococcus sanguinis with a sialyl galactose disaccharide bound | | Descriptor: | ACETATE ION, CALCIUM ION, N-glycolyl-alpha-neuraminic acid-(2-3)-methyl beta-D-galactopyranoside, ... | | Authors: | Loukachevitch, L.V, McCulloch, K.M, Vann, K.R, Wawrzak, Z, Anderson, S, Iverson, T.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Sialoglycan Binding by the Streptococcus sanguinis SrpA Adhesin.
J.Biol.Chem., 291, 2016
|
|
