7CAM
 
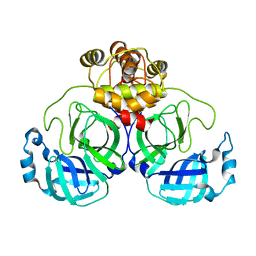 | | SARS-CoV-2 main protease (Mpro) apo structure (space group P212121) | | Descriptor: | 3C-like proteinase | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y. | | Deposit date: | 2020-06-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7CB7
 
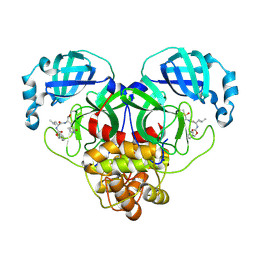 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
7WQA
 
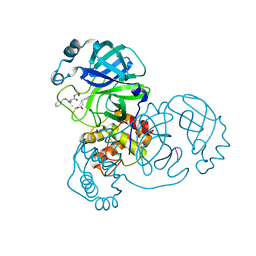 | | SARS-CoV-2 main protease in complex with Z-VAD-FMK | | Descriptor: | 3C-like proteinase, Z-VAD(OMe)-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SARS-CoV-2 main protease in complex with Z-VAD-FMK
To Be Published
|
|
7WQ9
 
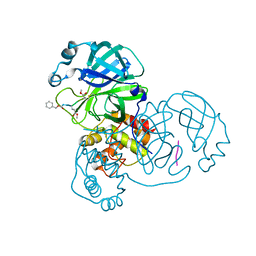 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK | | Descriptor: | 3C-like proteinase, Z-IETD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-IETD-FMK
To Be Published
|
|
7WQ8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK | | Descriptor: | 3C-like proteinase, Z-DEVD-FMK | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMK
To Be Published
|
|
7WQK
 
 | | wild-type SARS-CoV-2 main protease in complex with MG-132 | | Descriptor: | 3C-like proteinase, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | wild-type SARS-CoV-2 main protease in complex with MG-132
To Be Published
|
|
7WQB
 
 | | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chen, Y. | | Deposit date: | 2022-01-25 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | SARS-CoV-2 main protease mutant (P168A) in complex with MG-132
To Be Published
|
|
5HTL
 
 | | Structure of MshE with cdg | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MSHA biogenesis protein MshE | | Authors: | Chin, K.H, Wang, Y.C. | | Deposit date: | 2016-01-27 | | Release date: | 2016-10-05 | | Last modified: | 2016-10-12 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain.
Nat Commun, 7, 2016
|
|
4KGB
 
 | |
7XJW
 
 | | Crystal structure of canine coronavirus main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, ORF1a polyprotein | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chiu, Y.F, Chen, Y. | | Deposit date: | 2022-04-18 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Structural Comparison of SARS-CoV-2 Main Protease and Animal Coronaviral Main Protease Reveals Species-Specific Ligand Binding and Dimerization Mechanism.
Int J Mol Sci, 23, 2022
|
|
7DDC
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with Tafenoquine | | Descriptor: | 3C-like proteinase, Tafenoquine | | Authors: | Chen, Y, Wang, Y.C, Yang, C.S, Hung, M.C. | | Deposit date: | 2020-10-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Tafenoquine and its derivatives as inhibitors for the severe acute respiratory syndrome coronavirus 2.
J.Biol.Chem., 298, 2022
|
|
7DD0
 
 | |
7YA2
 
 | |
7X4Q
 
 | | LpCdnE UTP Mg complex | | Descriptor: | Cyclic dipyrimidine nucleotide synthase, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4A
 
 | | Native CD-NTase ClCdnE | | Descriptor: | ClCdnE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4P
 
 | | CD-NTase EfCdnE in complex with intermediate pppUpU | | Descriptor: | EfCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4C
 
 | | Native CD-NTase EfCdnE | | Descriptor: | EfCdnE, L(+)-TARTARIC ACID | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4F
 
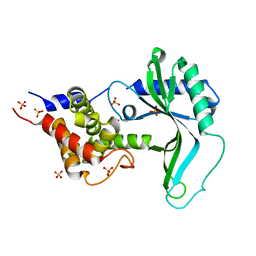 | | Native CD-NTase LpCdnE | | Descriptor: | Cyclic dipyrimidine nucleotide synthase, SULFATE ION | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4T
 
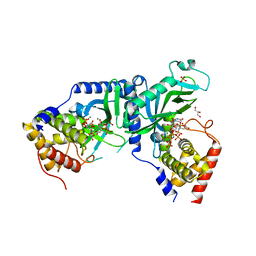 | | LpCdnE UMPNPP Mg complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, Cyclic dipyrimidine nucleotide synthase, GLYCEROL, ... | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
7X4G
 
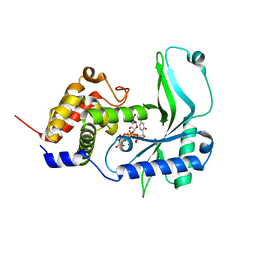 | | CD-NTase ClCdnE in complex with substrate UTP | | Descriptor: | ClCdnE, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Chen, Y, Ko, T.P, Yang, C.S, Wang, Y.C, Hou, M.H. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and functional implications of cyclic di-pyrimidine-synthesizing cGAS/DncV-like nucleotidyltransferases.
Nat Commun, 14, 2023
|
|
4WT2
 
 | | Co-crystal Structure of MDM2 in Complex with AM-7209 | | Descriptor: | 4-({[(3R,5R,6S)-1-[(1S)-2-(tert-butylsulfonyl)-1-cyclopropylethyl]-6-(4-chloro-3-fluorophenyl)-5-(3-chlorophenyl)-3-methyl-2-oxopiperidin-3-yl]acetyl}amino)-2-methoxybenzoic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shaffer, P.L, Huang, X, Yakowec, P, Long, A.M. | | Deposit date: | 2014-10-30 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Discovery of AM-7209, a Potent and Selective 4-Amidobenzoic Acid Inhibitor of the MDM2-p53 Interaction.
J.Med.Chem., 57, 2014
|
|
8HWJ
 
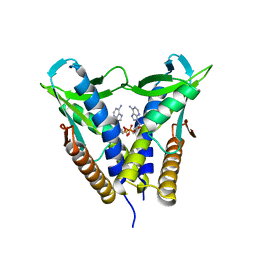 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWI
 
 | | Bacterial STING from Larkinella arboricola in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HY9
 
 | | Bacterial STING from Riemerella anatipestifer in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.462 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HY8
 
 | | Bacterial STING from Epilithonimonas lactis | | Descriptor: | CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
