4JYV
 
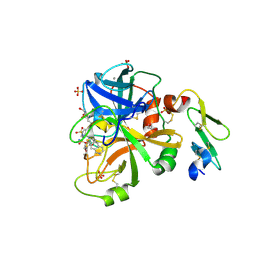 | |
229L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GUANIDINE, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-26 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
2MHM
 
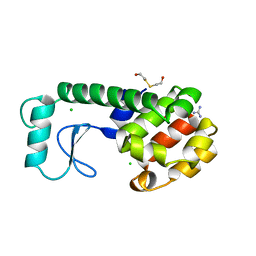
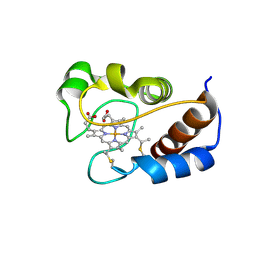 | | Solution structure of cytochrome c Y67H | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W.X, Wang, Z.H, Yang, Z.Z, Ying, T.L, Wu, H.M, Tan, X.S, Cao, C.Y, Huang, Z.X. | | Deposit date: | 2013-11-26 | | Release date: | 2014-10-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Cytochrome c Y67H Mutant to Function as a Peroxidase
Plos One, 9, 2014
|
|
2P8S
 
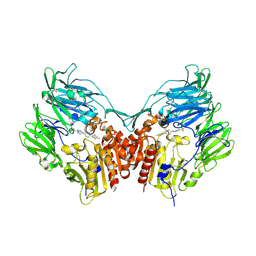 | | Human dipeptidyl peptidase IV/CD26 in complex with a cyclohexalamine inhibitor | | Descriptor: | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a novel, potent, and orally bioavailable cyclohexylamine DPP-4 inhibitor by application of molecular modeling and X-ray crystallography of sitagliptin
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2JUT
 
 | |
2JSY
 
 | | Solution structure of Tpx in the oxidized state | | Descriptor: | Probable thiol peroxidase | | Authors: | Jin, C, Lu, J. | | Deposit date: | 2007-07-17 | | Release date: | 2008-07-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Reversible conformational switch revealed by the redox structures of Bacillus subtilis thiol peroxidase
Biochem.Biophys.Res.Commun., 373, 2008
|
|
2JUR
 
 | |
2JUQ
 
 | |
7BQT
 
 | | Epstein-Barr virus, C12 portal dodecamer | | Descriptor: | Portal protein | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BTO
 
 | | EcoR124I-ArdA in the Translocation State | | Descriptor: | Antirestriction protein ArdA, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
4OC8
 
 | | DNA modification-dependent restriction endonuclease AspBHI | | Descriptor: | PHOSPHATE ION, restriction endonuclease AspBHI | | Authors: | Horton, J.R. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI.
Sci Rep, 4, 2014
|
|
7BQX
 
 | | Epstein-Barr virus, C5 portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BR7
 
 | | Epstein-Barr virus, C1 portal-proximal penton vertex, CATC binding | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Li, Z, Yu, X. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | CryoEM structure of the tegumented capsid of Epstein-Barr virus.
Cell Res., 30, 2020
|
|
7BTP
 
 | | EcoR124I-Ocr in Restriction-Alleviation State | | Descriptor: | Overcome classical restriction gp0.3, Type I restriction enzyme EcoR124II M protein, Type I restriction enzyme R Protein, ... | | Authors: | Gao, Y, Gao, P. | | Deposit date: | 2020-04-02 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structural insights into assembly, operation and inhibition of a type I restriction-modification system.
Nat Microbiol, 5, 2020
|
|
1JJG
 
 | | Solution Structure of Myxoma Virus Protein M156R | | Descriptor: | M156R | | Authors: | Ramelot, T.A, Cort, J.R, Yee, A.A, Arrowsmith, C.H, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-07-05 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Myxoma virus immunomodulatory protein M156R is a structural mimic of eukaryotic translation initiation factor eIF2alpha.
J.Mol.Biol., 322, 2002
|
|
6KWD
 
 | | Crystal Structure Analysis of Endo-beta-1,4-Xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
1PFN
 
 | | PF4-M2 CHIMERIC MUTANT WITH THE FIRST 10 N-TERMINAL RESIDUES OF R-PF4 REPLACED BY THE N-TERMINAL RESIDUES OF THE IL8 SEQUENCE. MODELS 16-27 OF A 27-MODEL SET. | | Descriptor: | PF4-M2 CHIMERA | | Authors: | Mayo, K.H, Roongta, V, Ilyina, E, Milius, R, Barker, S, Quinlan, C, La Rosa, G, Daly, T.J. | | Deposit date: | 1995-07-18 | | Release date: | 1996-01-29 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the 32-kDa platelet factor 4 ELR-motif N-terminal chimera: a symmetric tetramer.
Biochemistry, 34, 1995
|
|
6L1T
 
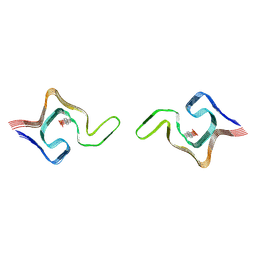 | | Cryo-EM structure of phosphorylated Tyr39 a-synuclein amyloid fibril | | Descriptor: | Alpha-synuclein | | Authors: | Liu, C, Li, Y.M, Zhao, K, Lim, Y.J, Liu, Z.Y. | | Deposit date: | 2019-09-30 | | Release date: | 2020-08-12 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Parkinson's disease-related phosphorylation at Tyr39 rearranges alpha-synuclein amyloid fibril structure revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5YF4
 
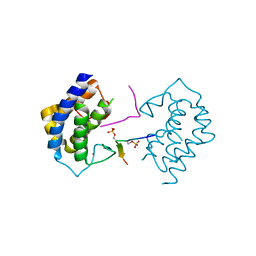 | | A kinase complex MST4-MOB4 | | Descriptor: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | Authors: | Chen, M, Zhou, Z.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
1O3Q
 
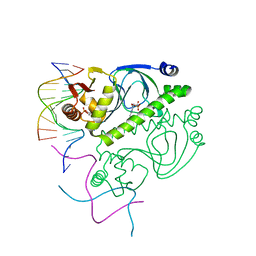 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*TP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*AP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
1O3R
 
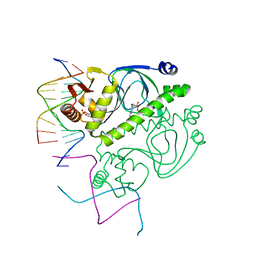 | | PROTEIN-DNA RECOGNITION AND DNA DEFORMATION REVEALED IN CRYSTAL STRUCTURES OF CAP-DNA COMPLEXES | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*T)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Chen, S, Vojtechovsky, J, Parkinson, G.N, Ebright, R.H, Berman, H.M. | | Deposit date: | 2003-03-18 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Indirect Readout of DNA Sequence at the Primary-kink Site in the CAP-DNA Complex: DNA Binding Specificity Based on Energetics of DNA Kinking
J.Mol.Biol., 314, 2001
|
|
6KW9
 
 | |
6KWF
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
6KWG
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-09-06 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
7W0S
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | Authors: | Zhang, H, Liang, X, Li, X.Z. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
