7D4K
 
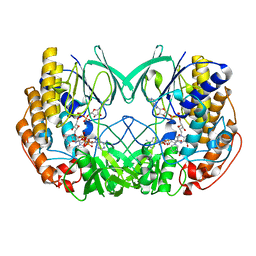 | | Crystal structure of Tmm from Pelagibacter sp. strain HTCC7211 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7EO0
 
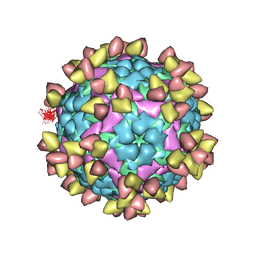 | | FOOT AND MOUTH DISEASE VIRUS O/TIBET/99-BOUND THE SINGLE CHAIN FRAGMEN ANTIBODY C4 | | Descriptor: | Ig heavy chain variable region, Ig lamda chain variable region, O/TIBET/99 VP1, ... | | Authors: | He, Y, Li, K. | | Deposit date: | 2021-04-21 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Two Cross-Protective Antigen Sites on Foot-and-Mouth Disease Virus Serotype O Structurally Revealed by Broadly Neutralizing Antibodies from Cattle.
J.Virol., 95, 2021
|
|
7V4Q
 
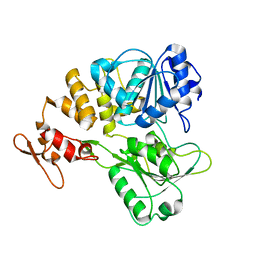 | |
7V4R
 
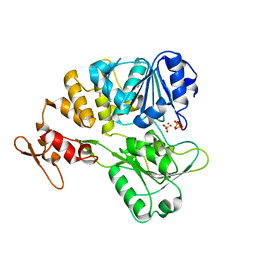 | | The crystal structure of KFDV NS3H bound with Pi | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, Serine protease NS3 | | Authors: | Zhang, C.Y, Jin, T.C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kyasanur Forest disease virus NS3 helicase: Insights into structure, activity, and inhibitors.
Int.J.Biol.Macromol., 2023
|
|
7XG7
 
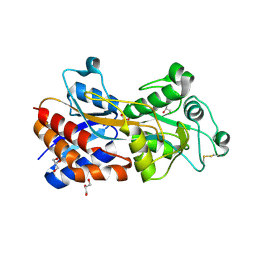 | | Crystal structure of PstS protein from cyanophage Syn19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Phosphate transporter subunit | | Authors: | Cai, K, Jiang, Y.L. | | Deposit date: | 2022-04-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of the cyanophage-encoded phosphate-binding protein: implications for enhanced phosphate uptake of infected cyanobacteria
Environ.Microbiol., 24, 2022
|
|
7XG8
 
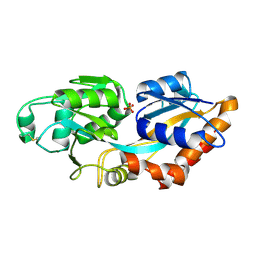 | | Crystal structure of PstS protein from cyanophage P-SSM2 | | Descriptor: | ABC transporter, substrate binding protein, phosphate, ... | | Authors: | Cai, K, Jiang, Y.L. | | Deposit date: | 2022-04-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Biochemical and structural characterization of the cyanophage-encoded phosphate-binding protein: implications for enhanced phosphate uptake of infected cyanobacteria
Environ.Microbiol., 24, 2022
|
|
5XM8
 
 | |
5YAT
 
 | |
5YNO
 
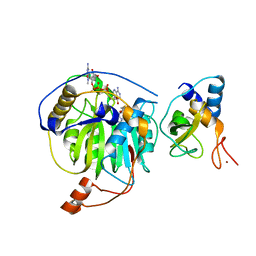 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5WXY
 
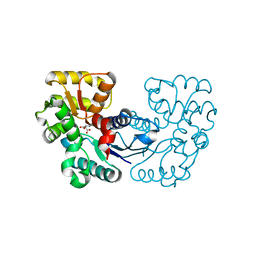 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
7DMX
 
 | |
5YNN
 
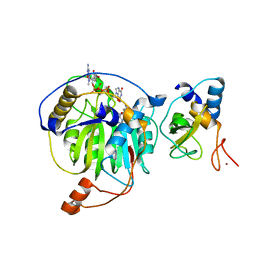 | | Crystal structure of MERS-CoV nsp16/nsp10complex bound to sinefungin and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5WXX
 
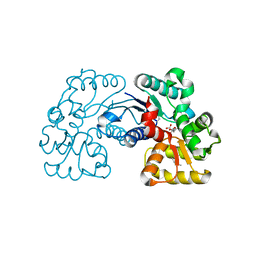 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with citrate | | Descriptor: | CITRIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5YNQ
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAH and m7GpppG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YN5
 
 | | Crystal structure of MERS-CoV nsp10/nsp16 complex | | Descriptor: | ZINC ION, nsp10 protein, nsp16 protein | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
7DNA
 
 | |
5YNP
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to sinefungin and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, SINEFUNGIN, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
5YNM
 
 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to SAM and m7GpppA | | Descriptor: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, ZINC ION, ... | | Authors: | Wei, S.M, Yang, L, Ke, Z.H, Guo, D.Y, Fan, C.P. | | Deposit date: | 2017-10-24 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
7F2B
 
 | |
5XM9
 
 | |
7DNB
 
 | | Crystal structure of PhoCl barrel | | Descriptor: | PhoCl Barrel, SODIUM ION | | Authors: | Wen, Y, Lemieux, J.M. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Photocleavable proteins that undergo fast and efficient dissociation.
Chem Sci, 12, 2021
|
|
5XMA
 
 | |
