4CDX
 
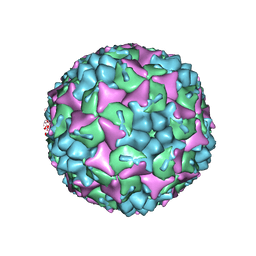 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
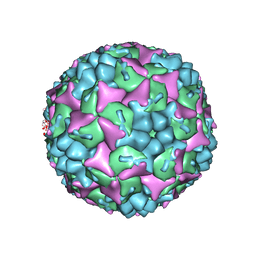 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDQ
 
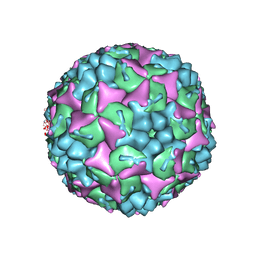 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEW
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
118D
 
 | |
7P5R
 
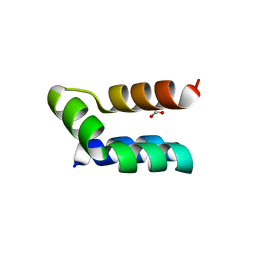 | | Racemic protein crystal structure of lacticin Q from Lactococcus lactis | | Descriptor: | D-Lacticin Q, FORMIC ACID, Lacticin Q | | Authors: | Lander, A.J, Li, X, Baumann, P, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Commun Chem, 6, 2023
|
|
3AQQ
 
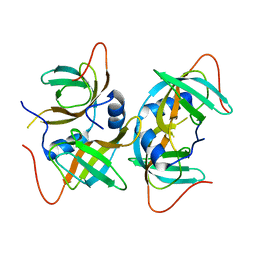 | | Crystal structure of human CRHSP-24 | | Descriptor: | Calcium-regulated heat stable protein 1 | | Authors: | Hou, H, Wang, F, Zhang, W, Wang, D, Li, X, Bartlam, M, Yao, X, Rao, Z. | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | CRHSP-24 is a novel cargo adaptor trafficking between stress granules and processing bodies
To be Published
|
|
8DKE
 
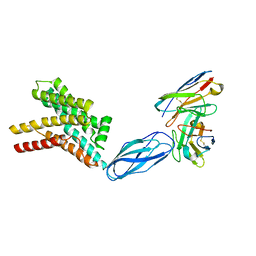 | |
8DKW
 
 | |
8DKI
 
 | |
8DKX
 
 | |
8DKM
 
 | |
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
3E73
 
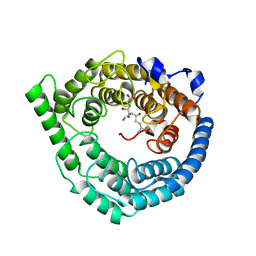 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3E6U
 
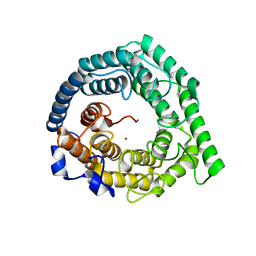 | | Crystal structure of Human LanCL1 | | Descriptor: | LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3ZIF
 
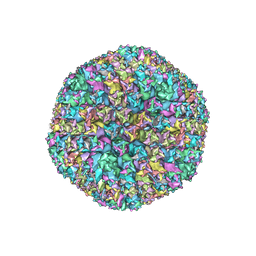 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
2R5W
 
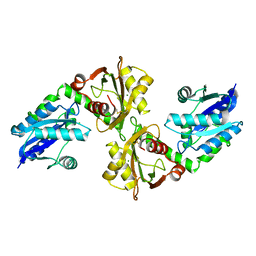 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
8DJM
 
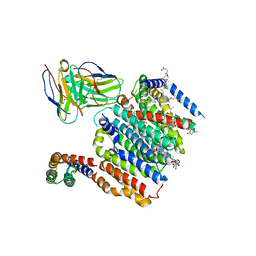 | | HMGCR-UBIAD1 Complex State 1 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-07-01 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
8DJK
 
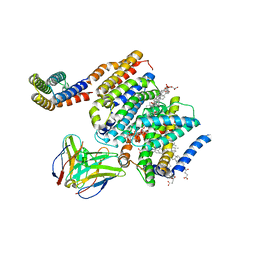 | | HMGCR-UBIAD1 Complex State 2 | | Descriptor: | 3-hydroxy-3-methylglutaryl-coenzyme A reductase, CHOLESTEROL HEMISUCCINATE, Digitonin, ... | | Authors: | Chen, H, Qi, X, Li, X. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Regulated degradation of HMG CoA reductase requires conformational changes in sterol-sensing domain.
Nat Commun, 13, 2022
|
|
7R8D
 
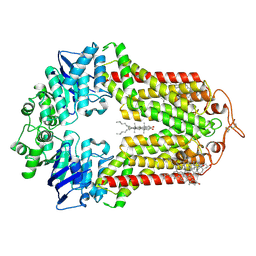 | | The structure of human ABCG1 E242Q with cholesterol | | Descriptor: | CHOLESTEROL, Isoform 4 of ATP-binding cassette sub-family G member 1 | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8A
 
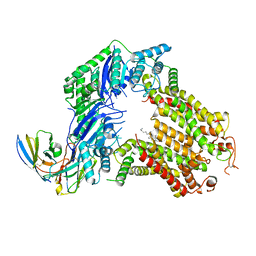 | | The structure of human ABCG5/ABCG8 purified from mammalian cells | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8B
 
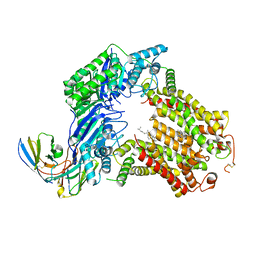 | | The structure of human ABCG5/ABCG8 supplemented with cholesterol | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R88
 
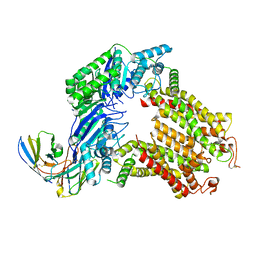 | | The structure of human ABCG5-I529W/ABCG8-WT | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R8E
 
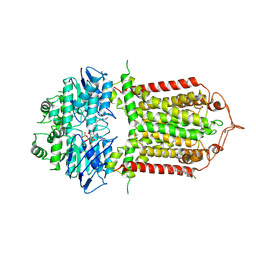 | | The structure of human ABCG1 E242Q complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL, Isoform 4 of ATP-binding cassette sub-family G member 1, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7R89
 
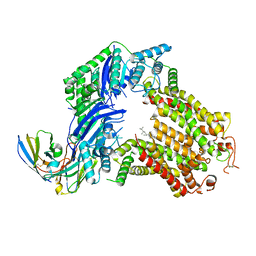 | | The structure of human ABCG5/ABCG8 purified from yeast | | Descriptor: | 2C7 Fab heavy chain, 2C7 Fab light chain, ATP-binding cassette sub-family G member 5, ... | | Authors: | Sun, Y, Li, X, Long, T. | | Deposit date: | 2021-06-26 | | Release date: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of cholesterol efflux via ABCG subfamily transporters.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
