5XS5
 
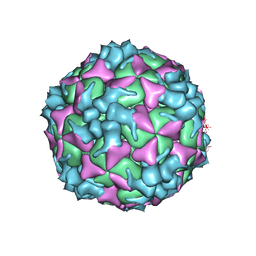 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5Z4Z
 
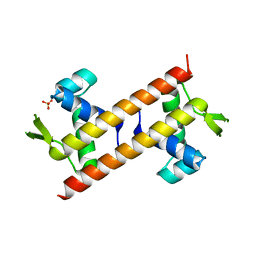 | |
5XS4
 
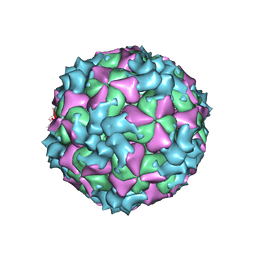 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5Z50
 
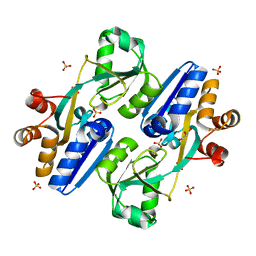 | | Crystal structure of PaCysB regulatory domain | | Descriptor: | Cys regulon transcriptional activator, GLYCEROL, SULFATE ION | | Authors: | Yang, C, Liang, H, Gan, J. | | Deposit date: | 2018-01-15 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa.
Mol.Microbiol., 111, 2019
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6JN2
 
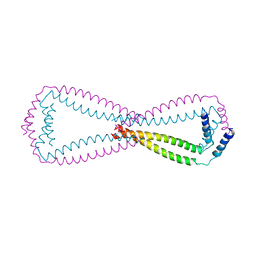 | | Crystal structure of the coiled-coil domains of human DOT1L in complex with AF10 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | Authors: | Song, X, Wang, M, Yang, N, Xu, R.M. | | Deposit date: | 2019-03-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A higher-order configuration of the heterodimeric DOT1L-AF10 coiled-coil domains potentiates their leukemogenenic activity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6U28
 
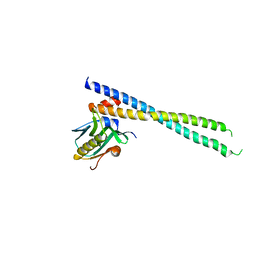 | | Crystal structure of 1918 NS1-ED W187A in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Cho, J.H, Zhao, B, Savage, N, Li, P. | | Deposit date: | 2019-08-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6BS2
 
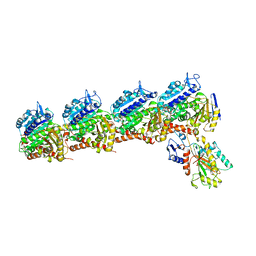 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 8b | | Descriptor: | 1-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
7XHF
 
 | |
7XHG
 
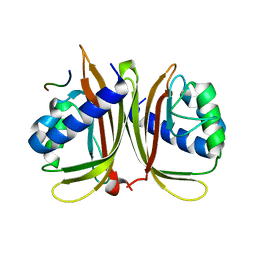 | |
6BR1
 
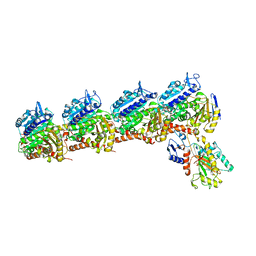 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BRF
 
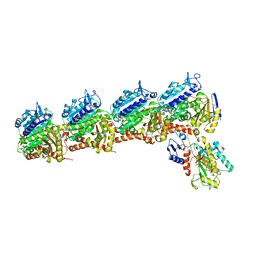 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[3,2-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BRY
 
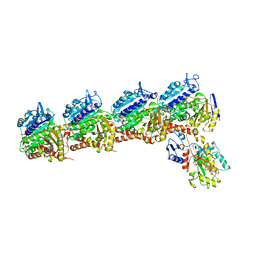 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
7ECW
 
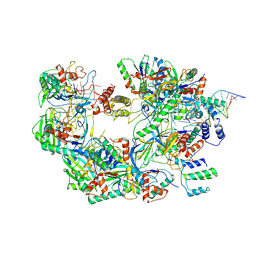 | | The Csy-AcrIF14-dsDNA complex | | Descriptor: | 54-MER DNA, AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7ECV
 
 | | The Csy-AcrIF14 complex | | Descriptor: | AcrIF14, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | Authors: | Zhang, L.X, Feng, Y. | | Deposit date: | 2021-03-13 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7DU0
 
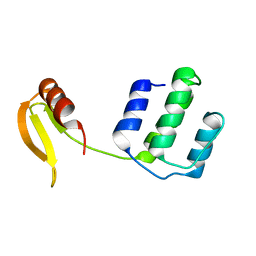 | | Structure of an type I-F anti-crispr protein | | Descriptor: | AcrIF14 | | Authors: | Teng, G, Yue, F. | | Deposit date: | 2021-01-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Insights into the dual functions of AcrIF14 during the inhibition of type I-F CRISPR-Cas surveillance complex.
Nucleic Acids Res., 49, 2021
|
|
7V3Z
 
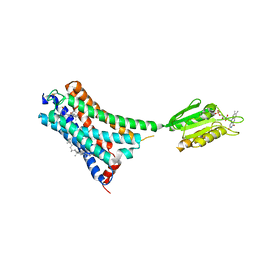 | | Structure of cannabinoid receptor type 1(CB1) | | Descriptor: | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, CHOLESTEROL, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Liu, Z.J, Shen, L, Hua, T, Yao, D.Q, Wu, L.J. | | Deposit date: | 2021-08-12 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | A Genetically Encoded F-19 NMR Probe Reveals the Allosteric Modulation Mechanism of Cannabinoid Receptor 1.
J.Am.Chem.Soc., 143, 2021
|
|
7WCM
 
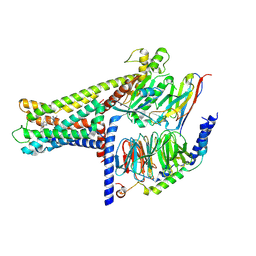 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist MBX-2982 | | Descriptor: | 2-[1-(5-ethylpyrimidin-2-yl)piperidin-4-yl]-4-[[4-(1,2,3,4-tetrazol-1-yl)phenoxy]methyl]-1,3-thiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
7WCN
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist AR231453 | | Descriptor: | Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
7DQ7
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 5F5 | | Descriptor: | 5F5 VH, 5F5 VL, Capsid protein VP4, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ4
 
 | | Cryo-EM structure of CAR triggered Coxsackievirus B1 A-particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPZ
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DPG
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle | | Descriptor: | VP2, VP3, Virion protein 1 | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q, Xia, N. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7DQ1
 
 | | Cryo-EM structure of Coxsackievirus B1 virion in complex with CAR at physiological temperature | | Descriptor: | Capsid protein VP4, Coxsackievirus and adenovirus receptor, VP2, ... | | Authors: | Li, S, Zhu, R, Xu, L, Cheng, T, Zheng, Q. | | Deposit date: | 2020-12-22 | | Release date: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
