6J5V
 
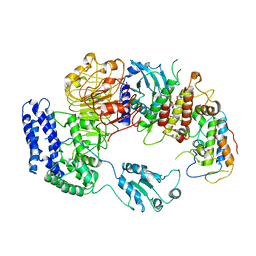 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | Descriptor: | Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, Protein kinase superfamily protein, ... | | Authors: | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
5NGH
 
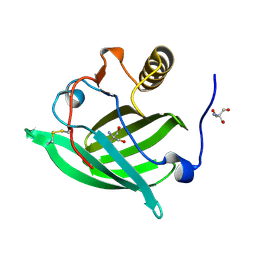 | |
5V8O
 
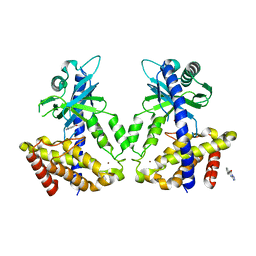 | | Discovery of a high affinity inhibitor of cGAS | | Descriptor: | 5-phenyltetrazolo[1,5-a]pyrimidin-7-ol, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Hall, J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay.
PLoS ONE, 12, 2017
|
|
4YVU
 
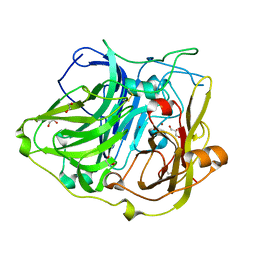 | | Crystal structure of CotA native enzyme in the acid condition, PH5.6 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4WXR
 
 | |
4WXP
 
 | |
3F9V
 
 | | Crystal Structure Of A Near Full-Length Archaeal MCM: Functional Insights For An AAA+ Hexameric Helicase | | Descriptor: | Minichromosome maintenance protein MCM | | Authors: | Chen, X.J, Brewster, A.S, Wang, G.G, Yu, X, Greenleaf, W, Tjajadi, M, Klein, M. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structure of a near-full-length archaeal MCM: Functional insights for an AAA+ hexameric helicase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4YVN
 
 | | Crystal structure of CotA laccase complexed with ABTS at a novel binding site | | Descriptor: | 1,2-ETHANEDIOL, 3-ETHYL-2-[(2Z)-2-(3-ETHYL-6-SULFO-1,3-BENZOTHIAZOL-2(3H)-YLIDENE)HYDRAZINO]-6-SULFO-3H-1,3-BENZOTHIAZOL-1-IUM, COPPER (II) ION, ... | | Authors: | Liu, Z.C, Xie, T, Wang, G.G. | | Deposit date: | 2015-03-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of CotA laccase complexed with 2,2-azinobis-(3-ethylbenzothiazoline-6-sulfonate) at a novel binding site
Acta Crystallogr.,Sect.F, 72, 2016
|
|
8ILQ
 
 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8I4T
 
 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
6A67
 
 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | Authors: | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | Deposit date: | 2018-06-26 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
7WZ7
 
 | | GPR110/G12 complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WXU
 
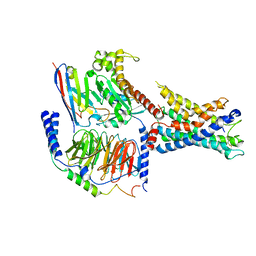 | | GPR110/Gq complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WY0
 
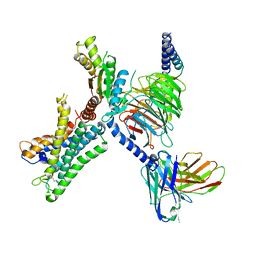 | | GPR110/G13 complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Engineered G alpha 13 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WXW
 
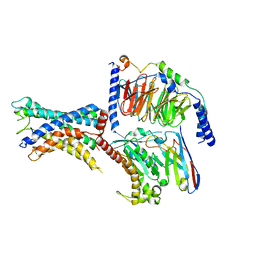 | | GPR110/Gs complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Engineered mini Galpha-s subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7X2V
 
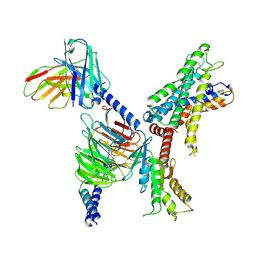 | | GPR110/Gi complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
8HCO
 
 | | Substrate-engaged TOM complex from yeast | | Descriptor: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | Authors: | Zhou, X.Y, Yang, Y.Q, Wang, G.P, Wang, S.S. | | Deposit date: | 2022-11-02 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5HU1
 
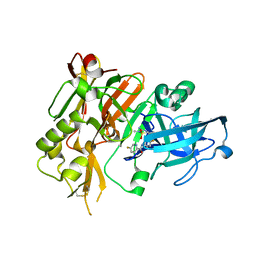 | | BACE1 in complex with (R)-N-(3-(3-amino-2,5-dimethyl-1,1-dioxido-5,6-dihydro-2H-1,2,4-thiadiazin-5-yl)-4-fluorophenyl)-5-fluoropicolinamide | | Descriptor: | Beta-secretase 1, N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxido-5,6-dihydro-2H-1,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
5DDT
 
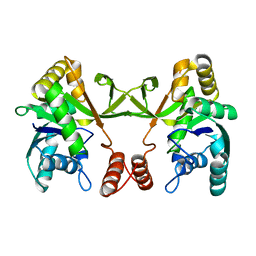 | |
8HFQ
 
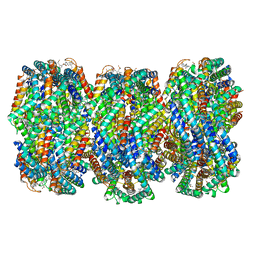 | | Cryo-EM structure of CpcL-PBS from cyanobacterium Synechocystis sp. PCC 6803 | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, Ferredoxin--NADP reductase, ... | | Authors: | Zheng, L, Zhang, Z, Wang, H, Zheng, Z, Gao, N, Zhao, J. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM and femtosecond spectroscopic studies provide mechanistic insight into the energy transfer in CpcL-phycobilisomes.
Nat Commun, 14, 2023
|
|
5HU0
 
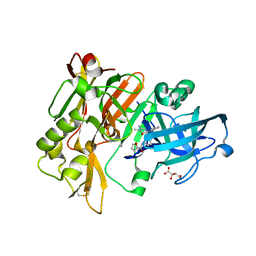 | | BACE1 in complex with 4-(3-(furan-2-carboxamido)phenyl)-1-methyl-5-oxo-4-phenylimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-{3-[(2E,4R)-2-imino-1-methyl-5-oxo-4-phenylimidazolidin-4-yl]phenyl}furan-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
5HTZ
 
 | | BACE1 in complex with (S)-5-(3-chloro-5-(5-(prop-1-yn-1-yl)pyridin-3-yl)thiophen-2-yl)-2,5-dimethyl-1,2,4-thiadiazinan-3-iminium 1,1-dioxide | | Descriptor: | (3E,5S)-5-{3-chloro-5-[5-(prop-1-yn-1-yl)pyridin-3-yl]thiophen-2-yl}-2,5-dimethyl-1,2,4-thiadiazinan-3-imine 1,1-dioxide, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
4OEW
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-iodo-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
5DDV
 
 | |
4OEX
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
