7U8R
 
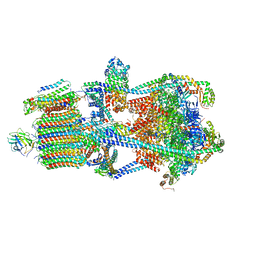 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7U8Q
 
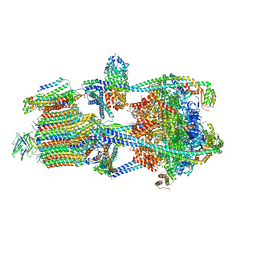 | | Structure of porcine kidney V-ATPase with SidK, Rotary State 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase H(+)-transporting lysosomal accessory protein 2, ATPase H+ transporting accessory protein 1, ... | | Authors: | Tan, Y.Z, Keon, K.A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM of endogenous mammalian V-ATPase interacting with the TLDc protein mEAK-7.
Life Sci Alliance, 5, 2022
|
|
7K9Z
 
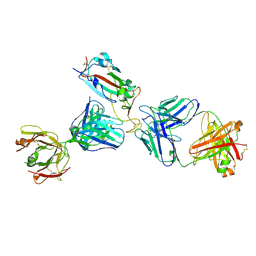 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | Authors: | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | Deposit date: | 2020-09-29 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
2KX4
 
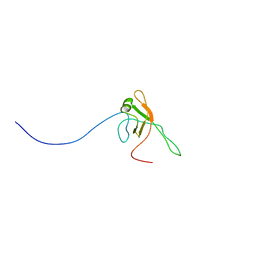 | | Solution structure of Bacteriophage Lambda gpFII | | Descriptor: | Tail attachment protein | | Authors: | Maxwell, K.L, Cardarelli, L, Neudecker, P, Davidson, A.R, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-04-26 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Phages have adapted the same protein fold to fulfill multiple functions in virion assembly.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JVO
 
 | | Crystal structure of bacteriophage HK97 gp6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Gp6 | | Authors: | Lam, R, Tuite, A, Battaile, K.P, Edwards, A.M, Maxwell, K.L, Chirgadze, N.Y. | | Deposit date: | 2009-09-17 | | Release date: | 2009-11-24 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of bacteriophage HK97 gp6: defining a large family of head-tail connector proteins.
J.Mol.Biol., 395, 2010
|
|
2LX4
 
 | |
6X8S
 
 | |
6X8Q
 
 | |
6X8P
 
 | |
6X8U
 
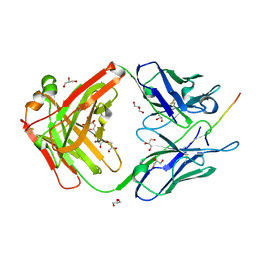 | |
9F9Y
 
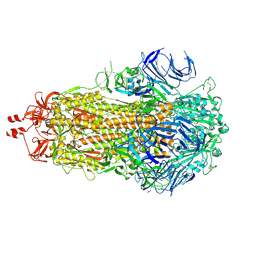 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
5BXH
 
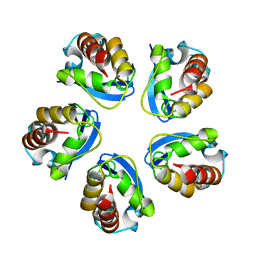 | |
5BXD
 
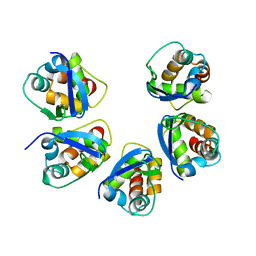 | |
5BXB
 
 | |
7UYL
 
 | | 850 Fab | | Descriptor: | 850 Fab Heavy Chain, 850 Fab Light Chain, GLYCEROL, ... | | Authors: | Kucharska, I, Prieto, K, Julien, J.P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire.
Plos Pathog., 18, 2022
|
|
7UYM
 
 | | 850 Fab in complex with NANPNANPNANP peptide | | Descriptor: | 850 Fab Heavy Chain, 850 Fab Light Chain, Circumsporozoite protein, ... | | Authors: | Kucharska, I, Prieto, K, Julien, J.P. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-density binding to Plasmodium falciparum circumsporozoite protein repeats by inhibitory antibody elicited in mouse with human immunoglobulin repertoire.
Plos Pathog., 18, 2022
|
|
6OJY
 
 | |
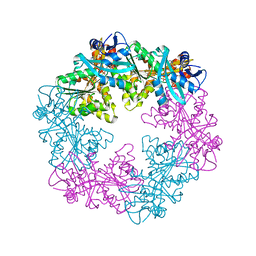
6OJX
 
 | |
6OKV
 
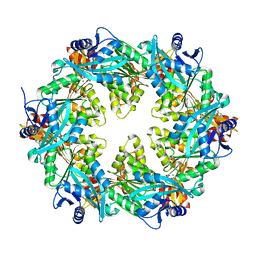 | |
6OLK
 
 | |
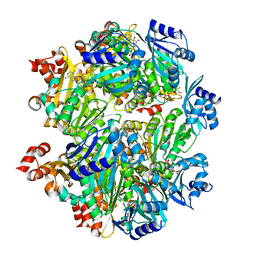
6OLM
 
 | |
5UFK
 
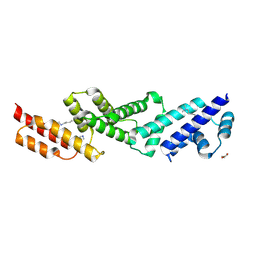 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-04 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5UF5
 
 | | Structure of the effector protein SidK (lpg0968) from Legionella pneumophila (domain-swapped dimer) | | Descriptor: | GLYCEROL, effector protein SidK | | Authors: | Beyrakhova, K, Xu, C, Boniecki, M.T, Cygler, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
5VKK
 
 | |
5VOX
 
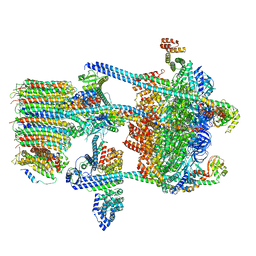 | | Yeast V-ATPase in complex with Legionella pneumophila effector SidK (rotational state 1) | | Descriptor: | V-type proton ATPase catalytic subunit A,V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Molecular basis for the binding and modulation of V-ATPase by a bacterial effector protein.
PLoS Pathog., 13, 2017
|
|
