3HEA
 
 | | The L29P/L124I mutation of Pseudomonas fluorescens esterase | | Descriptor: | Arylesterase, ETHYL ACETATE, GLYCEROL, ... | | Authors: | Kazlauskas, R.J, Schrag, J.D, Cheeseman, J.D, Morley, K.L. | | Deposit date: | 2009-05-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Switching catalysis from hydrolysis to perhydrolysis in Pseudomonas fluorescens esterase.
Biochemistry, 49, 2010
|
|
5ZU1
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*AP*GP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*CP*TP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-05 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.009 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5XLN
 
 | |
5ZUO
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
5ZUP
 
 | | Crystal Structure of BZ junction in diverse sequence | | Descriptor: | (5'-D(*AP*CP*GP*GP*TP*TP*TP*AP*TP*CP*GP*CP*GP*CP*GP*CP*G)-3'), (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*CP*AP*AP*TP*AP*AP*AP*CP*C)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Kim, K.K, Kim, D. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sequence preference and structural heterogeneity of BZ junctions.
Nucleic Acids Res., 46, 2018
|
|
6L4O
 
 | | Crystal structure of API5-FGF2 complex | | Descriptor: | Apoptosis inhibitor 5, Fibroblast growth factor 2 | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of mRNA export through API5 and nuclear FGF2 interaction.
Nucleic Acids Res., 48, 2020
|
|
6IMP
 
 | |
6K8W
 
 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6K8S
 
 | |
4EC2
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
6K8V
 
 | |
6K8T
 
 | |
6K8U
 
 | |
3OEQ
 
 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, with full length n-terminus | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3OER
 
 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | Descriptor: | COBALT (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
5X5T
 
 | |
5X5U
 
 | |
6B88
 
 | | E. coli LepB in complex with GNE0775 ((4S,7S,10S)-10-((S)-4-amino-2-(2-(4-(tert-butyl)phenyl)-4-methylpyrimidine-5-carboxamido)-N-methylbutanamido)-16,26-bis(2-aminoethoxy)-N-(2-iminoethyl)-7-methyl-6,9-dioxo-5,8-diaza-1,2(1,3)-dibenzenacyclodecaphane-4-carboxamide) | | Descriptor: | (8S,11S,14S)-14-{[(2S)-4-amino-2-{[2-(4-tert-butylphenyl)-4-methylpyrimidine-5-carbonyl]amino}butanoyl](methyl)amino}-3,18-bis(2-aminoethoxy)-N-[(2Z)-2-iminoethyl]-11-methyl-10,13-dioxo-9,12-diazatricyclo[13.3.1.1~2,6~]icosa-1(19),2(20),3,5,15,17-hexaene-8-carboxamide, PENTAETHYLENE GLYCOL, Signal peptidase I | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Optimized arylomycins are a new class of Gram-negative antibiotics.
Nature, 561, 2018
|
|
2FQL
 
 | |
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
2MSY
 
 | | Solution structure of Hox homeodomain | | Descriptor: | Homeobox protein Hox-C9 | | Authors: | Kim, H, Park, S, Han, J, Lee, B. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the interaction between the Hox and HMGB1 and understanding of the HMGB1-enhancing effect of Hox-DNA binding.
Biochim.Biophys.Acta, 1854, 2015
|
|
2LDM
 
 | |
4OZ5
 
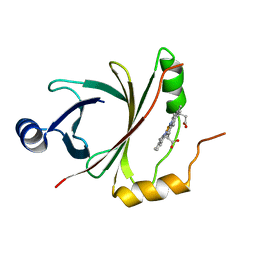 | | Bacillus subtilis HmoB | | Descriptor: | Bacillus subtilis HmoB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Choe, J, Park, S. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Bacillus subtilis HmoB
To Be Published
|
|
2LJ2
 
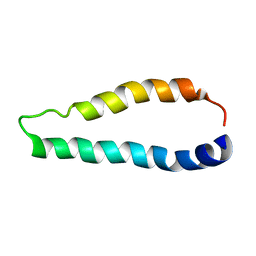 | | Integral membrane core domain of the mercury transporter MerF in lipid bilayer membranes | | Descriptor: | MerF | | Authors: | Das, B.B, Nothnagel, H.J, Lu, G.J, Son, W, Park, S, Tian, Y.B, Marassi, F.M, Opella, S.J. | | Deposit date: | 2011-09-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure determination of a membrane protein in proteoliposomes.
J.Am.Chem.Soc., 134, 2012
|
|
1QLQ
 
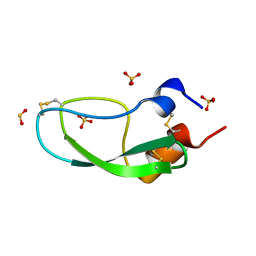 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
