7DTG
 
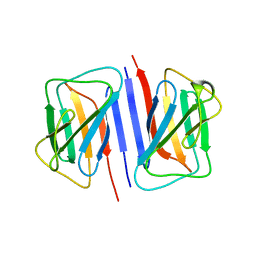 | |
7LKP
 
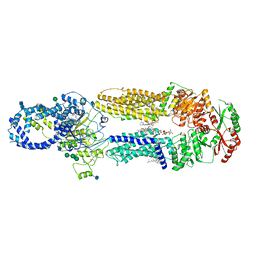 | | Structure of ATP-free human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-02 | | Release date: | 2021-03-03 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
7LKZ
 
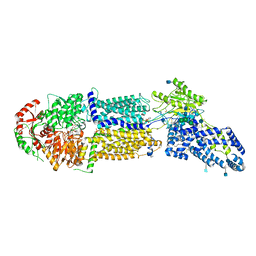 | | Structure of ATP-bound human ABCA4 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, F, Lee, J, Chen, J. | | Deposit date: | 2021-02-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Molecular structures of the eukaryotic retinal importer ABCA4.
Elife, 10, 2021
|
|
8ORD
 
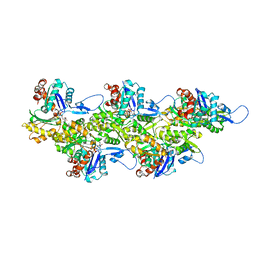 | | Cryo-EM map of zebrafish cardiac F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha 1b, ... | | Authors: | Bradshaw, M, Squire, J.M, Morris, E, Atkinson, G, Richardson, B, Lees, J, Paul, D.M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Zebrafish as a model for cardiac disease; Cryo-EM structure of native cardiac thin filaments from Danio Rerio.
J.Muscle Res.Cell.Motil., 44, 2023
|
|
1EZ3
 
 | |
5WBW
 
 | | Yeast Hsp104 fragment 1-360 | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-06-29 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase.
Biosci. Rep., 37, 2017
|
|
4UIP
 
 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6P79
 
 | | Engineered single chain antibody C9+C14 ScFv | | Descriptor: | Engineered antibody heavy chain, Engineered antibody light chain | | Authors: | Zhang, Y, Li, W, Marshall, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Computer-based Engineering of Thermostabilized Antibody Fragments.
Aiche J, 66, 2020
|
|
8K05
 
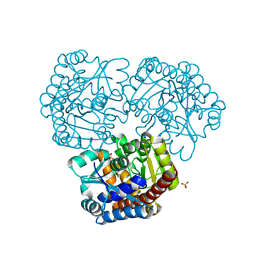 | |
8K07
 
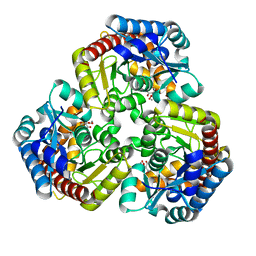 | |
8K06
 
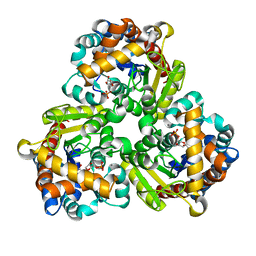 | | Pseudouridine 5'-monophosphate glycosylase from Arabidopsis thaliana -- PSU, R5P bound K185A mutant | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, MANGANESE (II) ION, PSEUDOURIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Lee, J.Y, Kim, S.H, Rhee, S.K. | | Deposit date: | 2023-07-07 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Structure and function of the pseudouridine 5'-monophosphate glycosylase PUMY from Arabidopsis thaliana.
Rna Biol., 21, 2024
|
|
7YLK
 
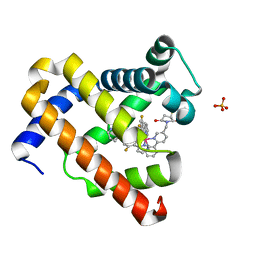 | | Myoglobin containing Ir complex | | Descriptor: | Myoglobin, SULFATE ION, delta-{1-([2,2'-bipyridin]-5-ylmethyl)pyrrolidine-2,5-dione}bis[2-(2,4-difluorophenyl)pyridine)]iridium(III), ... | | Authors: | Lee, J.H, Song, W.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Photocatalytic C-O Coupling Enzymes That Operate via Intramolecular Electron Transfer.
J.Am.Chem.Soc., 145, 2023
|
|
4J4L
 
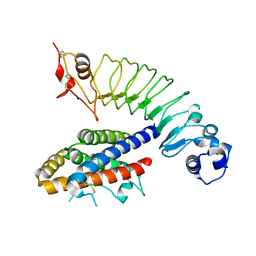 | |
6AMN
 
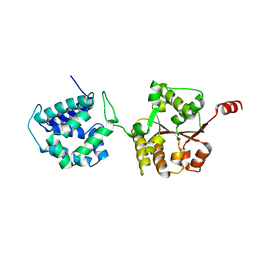 | | Crystal Structure of Hsp104 N Domain | | Descriptor: | Heat shock protein 104 | | Authors: | Lee, S. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.816 Å) | | Cite: | Overlapping and Specific Functions of the Hsp104 N Domain Define Its Role in Protein Disaggregation.
Sci Rep, 7, 2017
|
|
8U0T
 
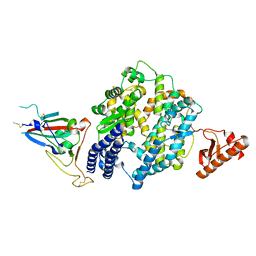 | |
3IX9
 
 | |
4KWC
 
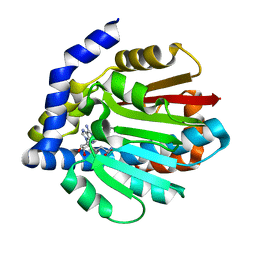 | |
4KVZ
 
 | |
2OXL
 
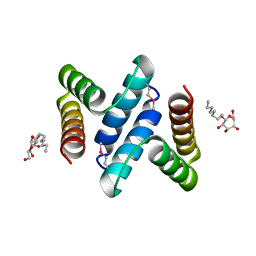 | | Structure and Function of the E. coli Protein YmgB: a Protein Critical for Biofilm Formation and Acid Resistance | | Descriptor: | Hypothetical protein ymgB, octyl beta-D-glucopyranoside | | Authors: | Page, R, Peti, W, Woods, T.K, Palermino, J.M, Doshi, O. | | Deposit date: | 2007-02-20 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Escherichia coli Protein YmgB: A Protein Critical for Biofilm Formation and Acid-resistance.
J.Mol.Biol., 373, 2007
|
|
2R5V
 
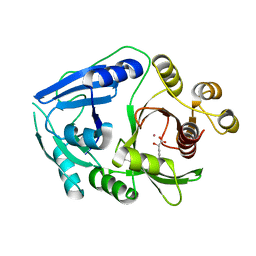 | | Hydroxymandelate Synthase Crystal Structure | | Descriptor: | (2S)-hydroxy(4-hydroxyphenyl)ethanoic acid, COBALT (II) ION, PCZA361.1, ... | | Authors: | Brownlee, J.M, He, P, Moran, G.R, Harrison, D.H.T. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate.
Biochemistry, 47, 2008
|
|
2QBR
 
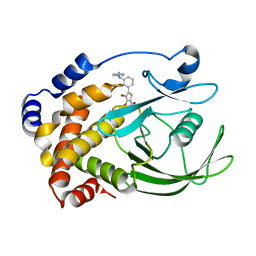 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-[3-(BENZYLAMINO)PHENYL]-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBQ
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-{3-[(3,3,5,5-TETRAMETHYLCYCLOHEXYL)AMINO]PHENYL}THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
2QBP
 
 | | Crystal structure of ptp1b-inhibitor complex | | Descriptor: | 5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W. | | Deposit date: | 2007-06-18 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based optimization of protein tyrosine phosphatase 1B inhibitors: from the active site to the second phosphotyrosine binding site.
J.Med.Chem., 50, 2007
|
|
7RFR
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-(4-methoxy-1H-indole-2-carbonyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Gajiwala, K.S, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFU
 
 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
