6JEY
 
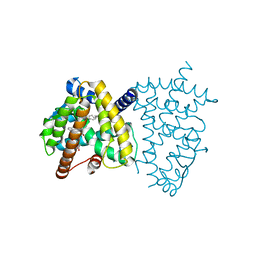 | | Covalent bond formation between ynone moiety of synthetic fatty acid and hPPARg-LBD | | Descriptor: | (9Z,12Z,15Z,18Z,21Z)-5-oxidanylidenetetracosa-9,12,15,18,21-pentaen-6-ynoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
6JF0
 
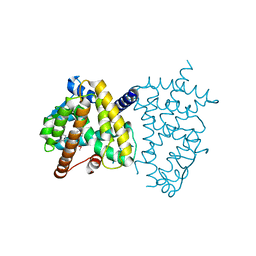 | |
6IQ5
 
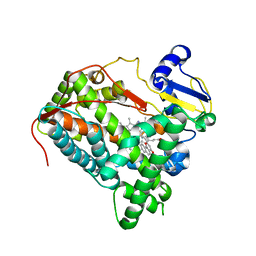 | | Crystal Structure of CYP1B1 and Inhibitor Having Azide Group | | Descriptor: | 2-(cis-4-azidocyclohexyl)-4H-naphtho[1,2-b]pyran-4-one, Cytochrome P450 1B1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kubo, M, Yamamoto, K, Itoh, T. | | Deposit date: | 2018-11-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Design and synthesis of selective CYP1B1 inhibitor via dearomatization of alpha-naphthoflavone.
Bioorg. Med. Chem., 27, 2019
|
|
6JEZ
 
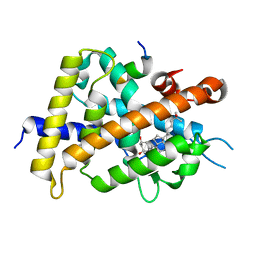 | | Covalent labeling of rVDR-LBD by turn-on fluorescent probe mediated by conjugate addition and cyclization | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, 7-(diethylamino)chromen-2-one, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kojima, H, Yamamoto, K, Itoh, T. | | Deposit date: | 2019-02-07 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclization Reaction-Based Turn-on Probe for Covalent Labeling of Target Proteins.
Cell Chem Biol, 27, 2020
|
|
5XPN
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5XPP
 
 | | Crystal structure of VDR-LBD complexed with 25RS-(Hydroxyphenyl)-2-methylidene-19,26,27-trinor-1,25-dihydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-oxidanyl-hexan-2-yl]-7~{ a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
3E9I
 
 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysine hydroxamate-AMP | | Descriptor: | 5'-O-{(R)-hydroxy[(L-lysylamino)oxy]phosphoryl}adenosine, Lysyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
3E9H
 
 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysylsulfamoyl adenosine | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysyl-tRNA synthetase, MAGNESIUM ION | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
5AZV
 
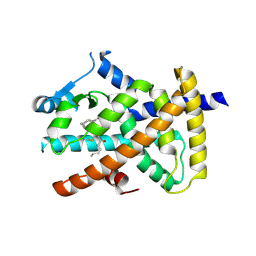 | | Crystal structure of hPPARgamma ligand binding domain complexed with 17-oxoDHA | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
5AZT
 
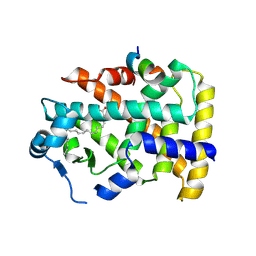 | | Ternary complex of hPPARalpha ligand binding domain, 17-oxoDHA and a SRC1 peptide | | Descriptor: | (4~{Z},7~{Z},10~{Z},13~{Z},19~{Z})-17-oxidanylidenedocosa-4,7,10,13,19-pentaenoic acid, 15-meric peptide from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Egawa, D, Itoh, T, Yamamoto, K. | | Deposit date: | 2015-10-23 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | 17-OxoDHA Is a PPAR alpha/gamma Dual Covalent Modifier and Agonist
Acs Chem.Biol., 2016
|
|
2D81
 
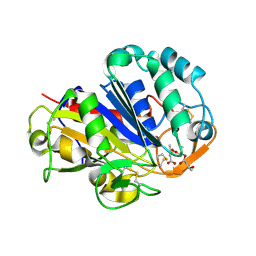 | | PHB depolymerase (S39A) complexed with R3HB trimer | | Descriptor: | (1R)-3-{[(1R)-3-METHOXY-1-METHYL-3-OXOPROPYL]OXY}-1-METHYL-3-OXOPROPYL (3R)-3-HYDROXYBUTANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, PHB depolymerase | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
2D80
 
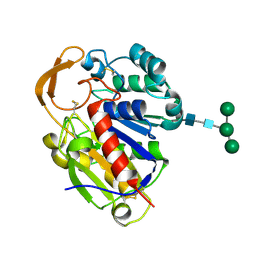 | | Crystal structure of PHB depolymerase from Penicillium funiculosum | | Descriptor: | PHB depolymerase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Hisano, T, Kasuya, K, Saito, T, Iwata, T, Miki, K. | | Deposit date: | 2005-11-30 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polyhydroxybutyrate Depolymerase from Penicillium funiculosum Provides Insights into the Recognition and Degradation of Biopolyesters
J.Mol.Biol., 356, 2006
|
|
7CUC
 
 | |
7D9W
 
 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCINE, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Putthapong, P, Hayashi, J, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9X
 
 | | Highly active mutant W525D of Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens | | Descriptor: | GAMMA-BUTYROLACTONE, GLYCEROL, GLYCINE, ... | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
7D9E
 
 | | Gamma-glutamyltranspeptidase from Pseudomonas nitroreducens complexed with L-DON | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, GLYCEROL, Gamma-glutamyltransferase 1 Threonine peptidase. MEROPS family T03 | | Authors: | Hibi, T, Sano, C, Itoh, T, Wakayama, M. | | Deposit date: | 2020-10-13 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mutagenesis and structure-based analysis of the role of Tryptophan525 of gamma-glutamyltranspeptidase from Pseudomonas nitroreducens.
Biochem.Biophys.Res.Commun., 534, 2021
|
|
3KSC
 
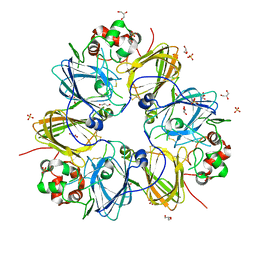 | | Crystal structure of pea prolegumin, an 11S seed globulin from Pisum sativum L. | | Descriptor: | GLYCEROL, LegA class, SULFATE ION | | Authors: | Tandang-Silvas, M.R.G, Fukuda, T, Fukuda, C, Prak, K, Cabanos, C, Kimura, A, Itoh, T, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-11-21 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
2H84
 
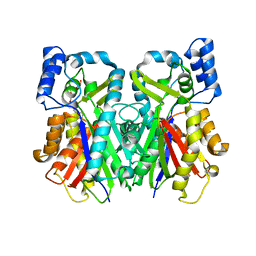 | | Crystal Structure of the C-terminal Type III Polyketide Synthase (PKS III) Domain of 'Steely1' (a Type I/III PKS Hybrid from Dictyostelium) | | Descriptor: | HEXAETHYLENE GLYCOL, Steely1 | | Authors: | Austin, M.B, Saito, T, Bowman, M.E, Haydock, S, Kato, A, Moore, B.S, Kay, R.R, Noel, J.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase.
Nat.Chem.Biol., 2, 2006
|
|
2E9Q
 
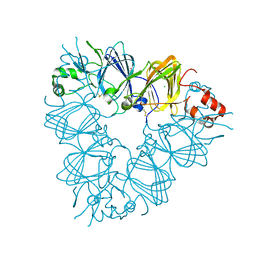 | | Recombinant pro-11S globulin of pumpkin | | Descriptor: | 11S globulin subunit beta, CHLORIDE ION, PHOSPHATE ION | | Authors: | Fukuda, T, Prak, K, Itoh, T, Masuda, T, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
3K3T
 
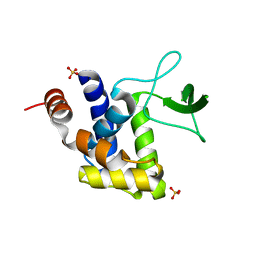 | | E185A mutant of peptidoglycan hydrolase from Sphingomonas sp. A1 | | Descriptor: | Peptidoglycan hydrolase FlgJ, SULFATE ION | | Authors: | Maruyama, Y, Ochiai, A, Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2009-10-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutational studies of the peptidoglycan hydrolase FlgJ of Sphingomonas sp. strain A1
J.Basic Microbiol., 50, 2010
|
|
7E2O
 
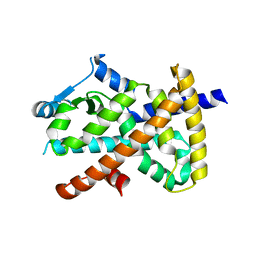 | | X-ray Crystal structure of PPARgamma R288H mutant. | | Descriptor: | Peroxisome proliferator-activated receptor gamma | | Authors: | Egawa, D, Itoh, T. | | Deposit date: | 2021-02-06 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Loss-of-Function R288H Mutant of Human PPAR gamma.
Biol.Pharm.Bull., 44, 2021
|
|
3VQU
 
 | | CRYSTAL STRUCTURE OF HUMAN MPS1 CATALYTIC DOMAIN IN COMPLEX WITH 4-[(4-amino-5-cyano-6-ethoxypyridin-2- yl)amino]benzamide | | Descriptor: | 4-[(4-amino-5-cyano-6-ethoxypyridin-2-yl)amino]benzamide, Dual specificity protein kinase TTK, IODIDE ION | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Itoh, T, Higashino, K, Okano, Y, Tadano, G, Tachibana, Y, Sato, Y, Inoue, M, Wada, T, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Tagashira, S, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Yamamoto, T, Higaki, M, Endoh, T, Ueda, K, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diaminopyridine-based potent and selective mps1 kinase inhibitors binding to an unusual flipped-Peptide conformation.
Acs Med.Chem.Lett., 3, 2012
|
|
7EFQ
 
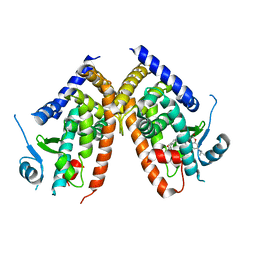 | | Crystal structure of hPPARgamma ligand binding domain complexed with rosiglitazone-based fluorescence probe | | Descriptor: | (5S)-5-[[4-[2-[[7-(diethylamino)-2-oxidanylidene-chromen-4-yl]-methyl-amino]ethoxy]phenyl]methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Yoshikawa, C, Ishida, H, Ohashi, N, Itoh, T. | | Deposit date: | 2021-03-23 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis of a Coumarin-Based PPAR gamma Fluorescence Probe for Competitive Binding Assay.
Int J Mol Sci, 22, 2021
|
|
3W1F
 
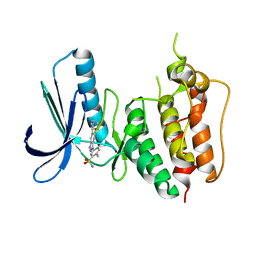 | | Crystal structure of Human MPS1 catalytic domain in complex with 5-(5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl)-2-methylbenzenesulfonamide | | Descriptor: | 5-[5-ethoxy-6-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-3-yl]-2-methylbenzenesulfonamide, Dual specificity protein kinase TTK | | Authors: | Kusakabe, K, Ide, N, Daigo, Y, Tachibana, Y, Itoh, T, Yamamoto, T, Hashizume, H, Hato, Y, Higashino, K, Okano, Y, Sato, Y, Inoue, M, Iguchi, M, Kanazawa, T, Ishioka, Y, Dohi, K, Kido, Y, Sakamoto, S, Yasuo, K, Maeda, M, Higaki, M, Ueda, K, Yoshizawa, H, Baba, Y, Shiota, T, Murai, H, Nakamura, Y. | | Deposit date: | 2012-11-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Indazole-based potent and cell-active Mps1 kinase inhibitors: rational design from pan-kinase inhibitor anthrapyrazolone (SP600125)
J.Med.Chem., 56, 2013
|
|
3VW4
 
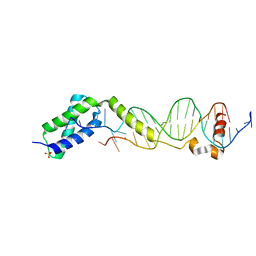 | | Crystal structure of the DNA-binding domain of ColE2-P9 Rep in complex with the replication origin | | Descriptor: | DNA (5'-D(P*AP*AP*TP*GP*AP*GP*AP*CP*CP*AP*GP*AP*TP*AP*AP*GP*CP*CP*TP*TP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*AP*AP*GP*GP*CP*TP*TP*AP*TP*CP*TP*GP*GP*TP*CP*TP*CP*AP*TP*T)-3'), Rep, ... | | Authors: | Itou, H, Yagura, M, Itoh, T, Shirakihara, Y. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Replication Origin Unwinding by An Initiator-Primase of Plasmid ColE2-P9: Duplex DNA Unwinding by A Single Protein
J.Biol.Chem., 290, 2015
|
|
