3BSA
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 2-({3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-(1,3-thiazol-5-yl)-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothia diazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase | | Authors: | Han, Q, Showalter, R.E, Zhao, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8GCR
 
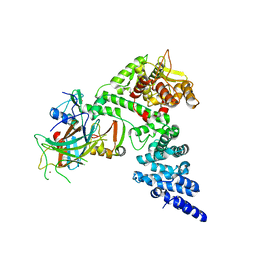 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
8HLW
 
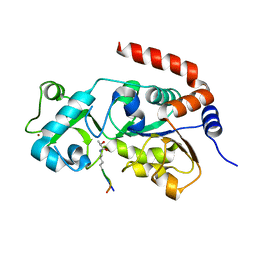 | | Crystal structure of SIRT3 in complex with H4K16la peptide | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Histone H4 residues 20-27, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Zhuming, F, Hao, Q. | | Deposit date: | 2022-12-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of SIRT3 as an eraser of H4K16la.
Iscience, 26, 2023
|
|
8HLY
 
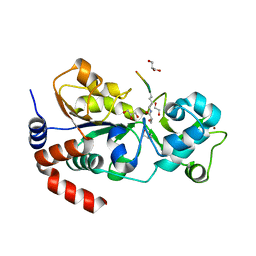 | |
4MBS
 
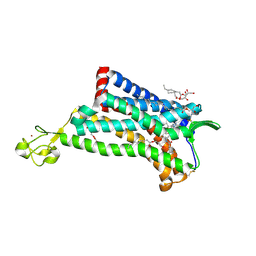 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
1GA5
 
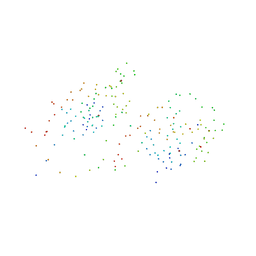 | | CRYSTAL STRUCTURE OF THE ORPHAN NUCLEAR RECEPTOR REV-ERB(ALPHA) DNA-BINDING DOMAIN BOUND TO ITS COGNATE RESPONSE ELEMENT | | Descriptor: | 5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*TP*AP*GP*GP*TP*CP*AP*G)-3', 5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*(5IT)P*G)-3', ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Sierk, M.L, Zhao, Q, Rastinejad, F. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | DNA Deformability as a Recognition Feature in the RevErb Response Element
Biochemistry, 40, 2001
|
|
1T0D
 
 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*(MTU)P*AP*GP*UP*CP*GP*CP*UP*GP*G)-3' | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1T0E
 
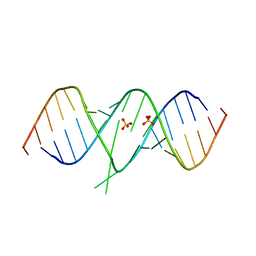 | | Crystal Structure of 2-aminopurine labelled bacterial decoding site RNA | | Descriptor: | 5'-R(*CP*GP*AP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*CP*C)-3', 5'-R(*GP*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*UP*CP*GP*G)-3', SULFATE ION | | Authors: | Shandrick, S, Zhao, Q, Han, Q, Ayida, B.K, Takahashi, M, Winters, G.C, Simonsen, K.B, Vourloumis, D, Hermann, T. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monitoring molecular recognition of the ribosomal decoding site.
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
1TIZ
 
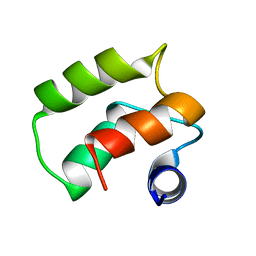 | | Solution Structure of a Calmodulin-Like Calcium-Binding Domain from Arabidopsis thaliana | | Descriptor: | calmodulin-related protein, putative | | Authors: | Song, J, Zhao, Q, Thao, S, Frederick, R.O, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: Solution structure of a calmodulin-like calcium-binding domain from Arabidopsis thaliana
J.Biomol.NMR, 30, 2004
|
|
1RJJ
 
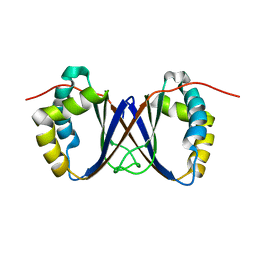 | | Solution structure of a homodimeric hypothetical protein, At5g22580, a structural genomics target from Arabidopsis thaliana | | Descriptor: | expressed protein | | Authors: | Cornilescu, G, Cornilescu, C.C, Zhao, Q, Frederick, R.O, Peterson, F.C, Thao, S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-11-19 | | Release date: | 2003-12-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a homodimeric hypothetical protein, at5g22580, a structural genomics target from Arabidopsis thaliana.
J.Biomol.Nmr, 29, 2004
|
|
9JJH
 
 | | Cryo-EM structure of a T=1 VLP of RHDV GI.2 with N-terminal 1-37 residues truncated | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol.
|
|
9JJJ
 
 | | Cryo-EM structure of a T=3 VLP of RHDV GI.2 | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol.
|
|
9JJG
 
 | | Cryo-EM structure of RHDV GI.2 virion | | Descriptor: | Genome polyprotein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol., 2014
|
|
9JJI
 
 | | Local refinement of RHDV GI.2 T=1 VLP | | Descriptor: | Capsid protein | | Authors: | Ruan, Z, Shao, Q, Song, Y, Hu, B, Fan, Z, Wei, H, Liu, Y, Wang, F, Fang, Q. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structures of RHDV reveal insights into capsid assembly and different conformations between mature virion and VLP
J.Virol.
|
|
1DSZ
 
 | | STRUCTURE OF THE RXR/RAR DNA-BINDING DOMAIN HETERODIMER IN COMPLEX WITH THE RETINOIC ACID RESPONSE ELEMENT DR1 | | Descriptor: | DNA (5'-D(*CP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*G)-3'), RETINOIC ACID RECEPTOR ALPHA, ... | | Authors: | Rastinejad, F, Wagner, T, Zhao, Q, Khorasanizadeh, S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1.
EMBO J., 19, 2000
|
|
6FK5
 
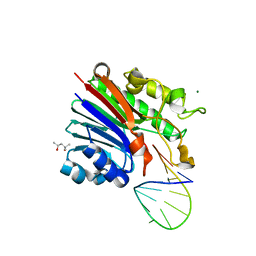 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to DNA substrate in presence of magnesium ion | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FKE
 
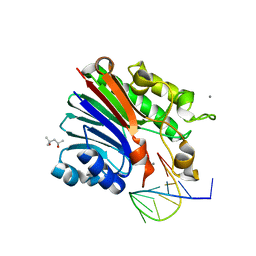 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to product DNA hairpin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*A)-3'), Exodeoxyribonuclease III, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FK4
 
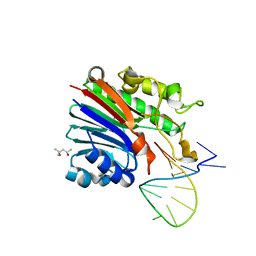 | | Structure of 3' phosphatase NExo (WT) from Neisseria bound to DNA substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), Exodeoxyribonuclease III | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
4XNW
 
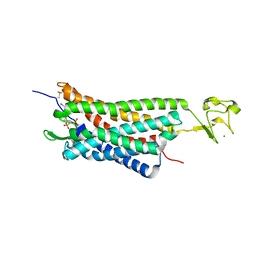 | | The human P2Y1 receptor in complex with MRS2500 | | Descriptor: | P2Y purinoceptor 1,Rubredoxin,P2Y purinoceptor 1, ZINC ION, [(1R,2S,4S,5S)-4-[2-iodo-6-(methylamino)-9H-purin-9-yl]-2-(phosphonooxy)bicyclo[3.1.0]hex-1-yl]methyl dihydrogen phosphate | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
4XNV
 
 | | The human P2Y1 receptor in complex with BPTU | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-[2-(2-tert-butylphenoxy)pyridin-3-yl]-3-[4-(trifluoromethoxy)phenyl]urea, CHOLESTEROL, ... | | Authors: | Zhang, D, Gao, Z, Jacobson, K, Han, G.W, Stevens, R, Zhao, Q, Wu, B, GPCR Network (GPCR) | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two disparate ligand-binding sites in the human P2Y1 receptor
Nature, 520, 2015
|
|
8JD2
 
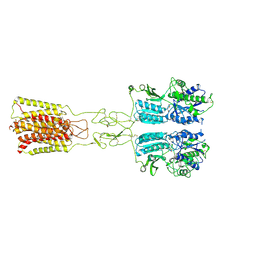 | | Cryo-EM structure of G protein-free mGlu2-mGlu3 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD1
 
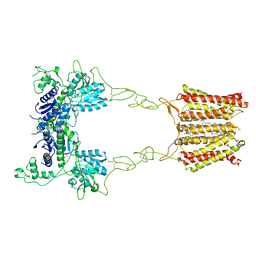 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in Rco state | | Descriptor: | CHOLESTEROL, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCW
 
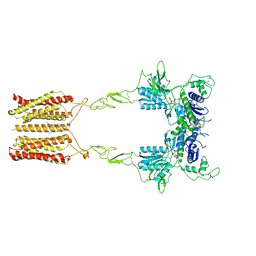 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD6
 
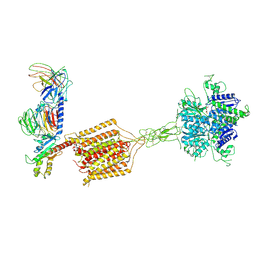 | | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCZ
 
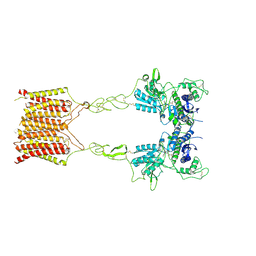 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
