6CHU
 
 | | CRYSTAL STRUCTURE OF PROTEIN CITE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM AND ACETATE | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2018-02-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1PRQ
 
 | | ACANTHAMOEBA CASTELLANII PROFILIN IA | | Descriptor: | PROFILIN IA | | Authors: | Fedorov, A.A, Pollard, T.D, Way, M, Lattman, E.E, Almo, S.C. | | Deposit date: | 1997-08-18 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal packing induces a conformational change in profilin-I from Acanthamoeba castellanii.
J.Struct.Biol., 123, 1998
|
|
4FX6
 
 | | Crystal structure of the mutant V182A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4GC4
 
 | | Crystal structure of the mutant R160A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4204 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4FXR
 
 | | Crystal structure of the mutant T159V.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-03 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4GK8
 
 | | Crystal structure of histidinol phosphate phosphatase (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN and L-histidinol arsenate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Histidinol-phosphatase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-08-10 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structural and Mechanistic Characterization of l-Histidinol Phosphate Phosphatase from the Polymerase and Histidinol Phosphatase Family of Proteins.
Biochemistry, 52, 2013
|
|
4GC3
 
 | | Crystal structure of L-HISTIDINOL PHOSPHATE PHOSPHATASE (HISK) from Lactococcus lactis subsp. lactis Il1403 complexed with ZN and sulfate | | Descriptor: | L-HISTIDINOL PHOSPHATE PHOSPHATASE, SULFATE ION, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Ghodge, S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2012-07-29 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and Mechanistic Characterization of l-Histidinol Phosphate Phosphatase from the Polymerase and Histidinol Phosphatase Family of Proteins.
Biochemistry, 52, 2013
|
|
4FX8
 
 | | Crystal structure of the mutant Q185A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9411 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1DFC
 
 | | CRYSTAL STRUCTURE OF HUMAN FASCIN, AN ACTIN-CROSSLINKING PROTEIN | | Descriptor: | FASCIN | | Authors: | Fedorov, A.A, Fedorov, E.V, Ono, S, Matsumura, F, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 1999-11-18 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein.
J.Mol.Biol., 400, 2010
|
|
3MTW
 
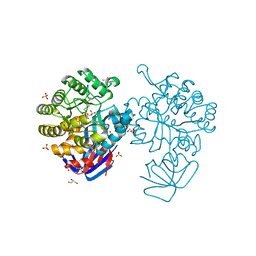 | | Crystal structure of L-Lysine, L-Arginine carboxypeptidase Cc2672 from Caulobacter Crescentus CB15 complexed with N-methyl phosphonate derivative of L-Arginine | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, L-Arginine carboxypeptidase Cc2672, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-05-01 | | Release date: | 2010-07-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Functional Identification and Structure Determination of Two Novel Prolidases from cog1228 in the Amidohydrolase Superfamily
Biochemistry, 49, 2010
|
|
1WUE
 
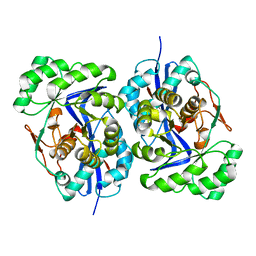 | | Crystal structure of protein GI:29375081, unknown member of enolase superfamily from enterococcus faecalis V583 | | Descriptor: | mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-05 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1WUF
 
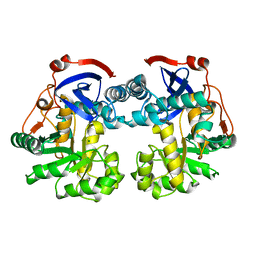 | | Crystal structure of protein GI:16801725, member of Enolase superfamily from Listeria innocua Clip11262 | | Descriptor: | MAGNESIUM ION, hypothetical protein lin2664 | | Authors: | Fedorov, A.A, Fedorov, E.V, Yew, W.S, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-12-07 | | Release date: | 2004-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1XM5
 
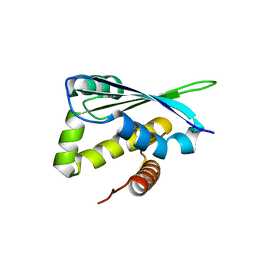 | | Crystal structure of metal-dependent hydrolase ybeY from E. coli, Pfam UPF0054 | | Descriptor: | Hypothetical UPF0054 protein ybeY, NICKEL (II) ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Shi, W, Ramagopal, U.A, Thirumuruhan, R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-10-01 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ybeY protein from Escherichia coli is a metalloprotein.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3JZU
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | Descriptor: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3KUM
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Arg-L-Tyr | | Descriptor: | ARGININE, Dipeptide Epimerase, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-11-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LI0
 
 | | Crystal structure of the mutant R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3LHT
 
 | | Crystal structure of the mutant V201F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LI1
 
 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LLD
 
 | | Crystal structure of the mutant S127G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LHU
 
 | | Crystal structure of the mutant I199F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LLF
 
 | | Crystal structure of the mutant S127P of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LHW
 
 | | Crystal structure of the mutant V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LHY
 
 | | Crystal structure of the mutant I199A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LTP
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-02-16 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LTY
 
 | | Crystal structure of the mutant V182A,I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-02-16 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
