6PIA
 
 | |
6PHR
 
 | |
6PI1
 
 | |
6PIC
 
 | |
6PZS
 
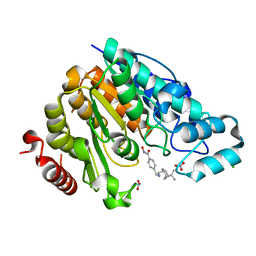 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with JR005 | | Descriptor: | 4-[({[(1R,2R,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl]methyl}amino)methyl]-N-hydroxybenzamide, CHLORIDE ION, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-08-01 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Exploring Structural Determinants of Inhibitor Affinity and Selectivity in Complexes with Histone Deacetylase 6.
J.Med.Chem., 63, 2020
|
|
6PYE
 
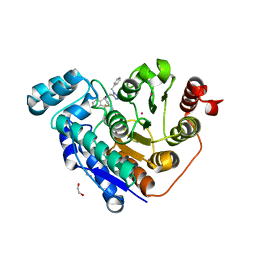 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NR160 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-[(1-benzyl-1H-tetrazol-5-yl)methyl]-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-2-(trifluoromethyl)benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-07-29 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.480003 Å) | | Cite: | Multicomponent Synthesis, Binding Mode, and Structure-Activity Relationship of Selective Histone Deacetylase 6 (HDAC6) Inhibitors with Bifurcated Capping Groups.
J.Med.Chem., 63, 2020
|
|
6PZR
 
 | |
6Q0Z
 
 | |
2FOQ
 
 | |
2FOU
 
 | | Human Carbonic Anhydrase II complexed with two-prong inhibitors | | Descriptor: | COPPER (II) ION, Carbonic Anhydrase II, GLYCEROL, ... | | Authors: | Jude, K.M, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
2FOV
 
 | | Human Carbonic Anhydrase II complexed with two-prong inhibitors | | Descriptor: | ACETONITRILE, COPPER (II) ION, Carbonic Anhydrase II, ... | | Authors: | Jude, K.M, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
6PZO
 
 | |
6PZU
 
 | |
2FOY
 
 | | Human Carbonic Anhydrase I complexed with a two-prong inhibitor | | Descriptor: | Carbonic anhydrase 1, ZINC ION, {2,2'-[(2-{[4-(AMINOSULFONYL)BENZOYL]AMINO}ETHYL)IMINO]DIACETATO(2-)-KAPPAO}COPPER | | Authors: | Jude, K.M, Banerjee, A.L, Haldar, M.K, Manokaran, S, Roy, B, Mallik, S, Srivastava, D.K, Christianson, D.W. | | Deposit date: | 2006-01-14 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
2FOS
 
 | | Human Carbonic Anhydrase II complexed with two-prong inhibitors | | Descriptor: | COPPER (II) ION, Carbonic Anhydrase II, ZINC ION, ... | | Authors: | Jude, K.M, Christianson, D.W. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ultrahigh resolution crystal structures of human carbonic anhydrases I and II complexed with two-prong inhibitors reveal the molecular basis of high affinity.
J.Am.Chem.Soc., 128, 2006
|
|
6OS6
 
 | |
6OFV
 
 | | F95Q Epi-isozizaene synthase | | Descriptor: | Epi-isozizaene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of F95Q epi-isozizaene synthase, an engineered sesquiterpene cyclase that generates biofuel precursors beta- and gamma-curcumene.
J.Struct.Biol., 207, 2019
|
|
6OS5
 
 | |
2GO3
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with imidazole. | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
2GO4
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with TU-514 | | Descriptor: | 1,5-ANHYDRO-2-C-(CARBOXYMETHYL-N-HYDROXYAMIDE)-2-DEOXY-3-O-MYRISTOYL-D-GLUCITOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
2E4O
 
 | | X-ray Crystal Structure of Aristolochene Synthase from Aspergillus terreus and the Evolution of Templates for the Cyclization of Farnesyl Diphosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Di Costanzo, L, Cane, D.E, Christianson, D.W. | | Deposit date: | 2006-12-15 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of aristolochene synthase from Aspergillus terreus and evolution of templates for the cyclization of farnesyl diphosphate.
Biochemistry, 46, 2007
|
|
2H4N
 
 | | H94N CARBONIC ANHYDRASE II COMPLEXED WITH ACETAZOLAMIDE | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine --> carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity.
Biochemistry, 36, 1997
|
|
2HG2
 
 | |
1YHC
 
 | | Crystal structure of Aquifex aeolicus LpxC deacetylase complexed with cacodylate | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hernick, M, Gennadios, H.A, Whittington, D.A, Rusche, K.M, Christianson, D.W, Fierke, C.A. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine Deacetylase Functions through a General Acid-Base Catalyst Pair Mechanism
J.Biol.Chem., 280, 2005
|
|
1YH8
 
 | | Crystal structure of Aquifex aeolicus LpxC deacetylase complexed with palmitate | | Descriptor: | CHLORIDE ION, PALMITOLEIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Hernick, M, Gennadios, H.A, Whittington, D.A, Rusche, K.M, Christianson, D.W, Fierke, C.A. | | Deposit date: | 2005-01-07 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine Deacetylase Functions through a General Acid-Base Catalyst Pair Mechanism
J.Biol.Chem., 280, 2005
|
|
