6JLS
 
 | | Crystal Structure of FMN-dependent Cysteine Decarboxylases TvaF from Thioviridamide Biosynthesis | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative flavoprotein decarboxylase | | Authors: | Li, J, Lu, J, Wang, H, Zhu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the FMN-Dependent Cysteine Decarboxylase from Thioviridamide Biosynthesis.
Org.Lett., 21, 2019
|
|
6CDN
 
 | | Crystal structure of cysteine-bound ferrous form of the crosslinked Cl-Tyr157 human cysteine dioxygenase | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
8IU2
 
 | | Cryo-EM structure of Long-wave-sensitive opsin 1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Peng, Q, Cheng, X.Y, Li, J, Lu, Q.Y, Li, Y.Y, Zhang, J. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of Long-wave-sensitive opsin 1
To Be Published
|
|
7VLQ
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P212121 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhang, J, Li, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.939106 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLO
 
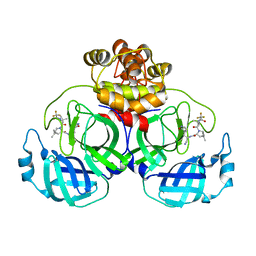 | | Crystal structure of SARS coronavirus main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.0227 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VLP
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with PF07321332 in spacegroup P1211 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, Replicase polyprotein 1a | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50251937 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
7VTC
 
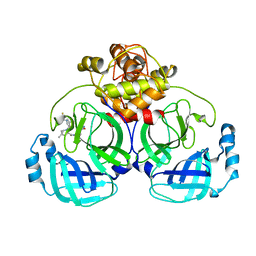 | | Crystal structure of MERS main protease in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Lin, C, Zhong, F.L, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53865623 Å) | | Cite: | Structural Basis of the Main Proteases of Coronavirus Bound to Drug Candidate PF-07321332.
J.Virol., 96, 2022
|
|
2JOH
 
 | |
5HC3
 
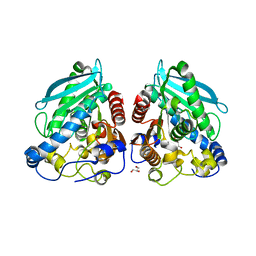 | | The structure of esterase Est22 | | Descriptor: | GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
5HC2
 
 | |
6DQ3
 
 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
6XLT
 
 | |
6XLS
 
 | |
6E87
 
 | |
6XMV
 
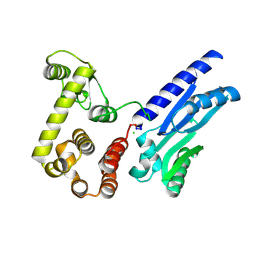 | | The crystal structure of Neisseria gonorrhoeae DolP (NGO1985) | | Descriptor: | CHLORIDE ION, Hemolysin,Endolysin | | Authors: | Li, J, Peterson Brown, L.E, Reed, R.W, Wierzbicki, I.H, Sikora, A.E, Korotkov, K.V. | | Deposit date: | 2020-06-30 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structure and function of a new component of Bam complex in Neisseria
To Be Published
|
|
6VHH
 
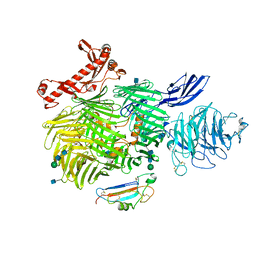 | | Human Teneurin-2 and human Latrophilin-3 binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G protein-coupled receptor L3, ... | | Authors: | Xie, Y, Li, J, Arac, D, Zhao, M. | | Deposit date: | 2020-01-09 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism.
Nat Commun, 11, 2020
|
|
6XLR
 
 | |
8U3P
 
 | |
6DKA
 
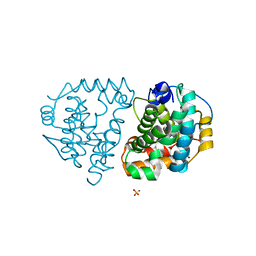 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | CYANAMIDE, DNA damage-inducible protein, SULFATE ION, ... | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
5HC5
 
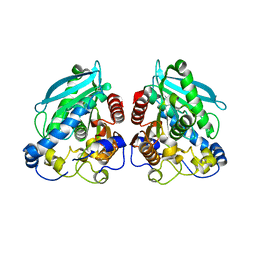 | |
7VFJ
 
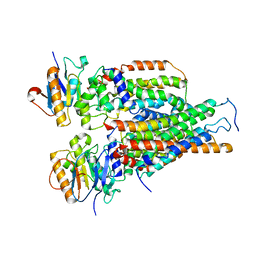 | | Cytochrome c-type biogenesis protein CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
6DK9
 
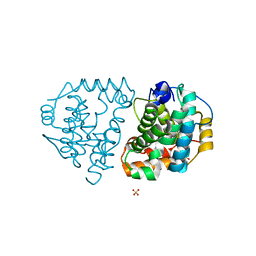 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
2FJ3
 
 | |
5YTK
 
 | | Crystal structure of SIRT3 bound to a leucylated AceCS2 | | Descriptor: | AceCS2-KLeu, LEUCINE, LYSINE, ... | | Authors: | Li, J, Gong, W, Xu, Y. | | Deposit date: | 2017-11-18 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations
Cell Metab., 27, 2018
|
|
5HC4
 
 | | Structure of esterase Est22 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Lipolytic enzyme | | Authors: | Li, J, Huang, J. | | Deposit date: | 2016-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights of a hormone sensitive lipase homologue Est22.
Sci Rep, 6, 2016
|
|
