2FUQ
 
 | | Crystal Structure of Heparinase II | | Descriptor: | FORMIC ACID, PHOSPHATE ION, ZINC ION, ... | | Authors: | Shaya, D, Cygler, M. | | Deposit date: | 2006-01-27 | | Release date: | 2006-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Heparinase II from Pedobacter heparinus and Its Complex with a Disaccharide Product.
J.Biol.Chem., 281, 2006
|
|
2GML
 
 | | Crystal Structure of Catalytic Domain of E.coli RluF | | Descriptor: | Ribosomal large subunit pseudouridine synthase F | | Authors: | Sunita, S, Zhenxing, H, Swaathi, J, Cygler, M, Matte, A, Sivaraman, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-07-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Organization and Crystal Structure of the Catalytic Domain of E.coli RluF, a Pseudouridine Synthase that Acts on 23S rRNA
J.Mol.Biol., 359, 2006
|
|
2FLO
 
 | |
2H7A
 
 | | NMR Structure of the Conserved Protein YcgL from Escherichia coli representing the DUF709 Family Reveals a Novel a/b/a Sandwich Fold | | Descriptor: | Hypothetical protein ycgL | | Authors: | Minailiuc, O.M, Vavelyuk, O, Ekiel, I, Hung, M.-Ni, Cygler, M, Gandhi, S, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-06-01 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of YcgL, a conserved protein from Escherichia coli representing the DUF709 family, with a novel alpha/beta/alpha sandwich fold.
Proteins, 66, 2007
|
|
2H8L
 
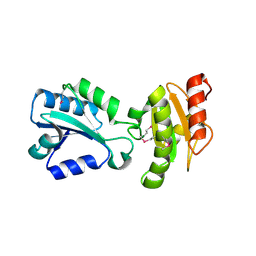 | |
2HXW
 
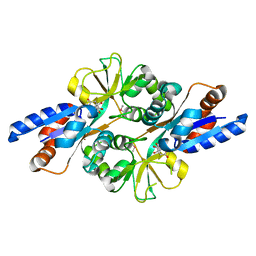 | | Crystal Structure of Peb3 from Campylobacter jejuni | | Descriptor: | CITRATE ANION, Major antigenic peptide PEB3 | | Authors: | Rangarajan, E.S, Bhatia, S, Watson, D.C, Munger, C, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-08-04 | | Release date: | 2007-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural context for protein N-glycosylation in bacteria: The structure of PEB3, an adhesin from Campylobacter jejuni.
Protein Sci., 16, 2007
|
|
1IJI
 
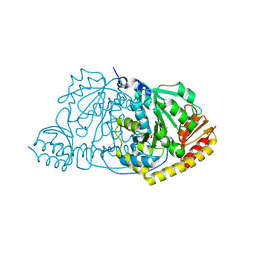 | | Crystal Structure of L-Histidinol Phosphate Aminotransferase with PLP | | Descriptor: | Histidinol Phosphate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Li, Y, Larocque, R, Schrag, J.D, Cygler, M, Matte, A. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-29 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1GZ0
 
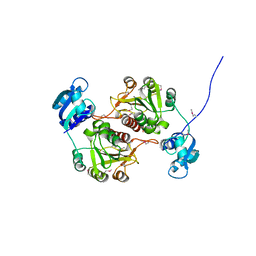 | | 23S RIBOSOMAL RNA G2251 2'O-METHYLTRANSFERASE RLMB | | Descriptor: | HYPOTHETICAL TRNA/RRNA METHYLTRANSFERASE YJFH | | Authors: | Michel, G, Cygler, M. | | Deposit date: | 2002-05-03 | | Release date: | 2002-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Rlmb 23S Rrna Methyltransferase Reveals a New Methyltransferase Fold with a Unique Knot
Structure, 10, 2002
|
|
1Q7L
 
 | | Zn-binding domain of the T347G mutant of human aminoacylase-I | | Descriptor: | Aminoacylase-1, GLYCINE, ZINC ION | | Authors: | Lindner, H.A, Lunin, V.V, Alary, A, Hecker, R, Cygler, M, Menard, R. | | Deposit date: | 2003-08-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family.
J.Biol.Chem., 278, 2003
|
|
1PTM
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, PHOSPHATE ION, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-23 | | Release date: | 2003-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1PVJ
 
 | | Crystal structure of the Streptococcal pyrogenic exotoxin B (SpeB)- inhibitor complex | | Descriptor: | (3R)-3-{[(BENZYLOXY)CARBONYL]AMINO}-2-OXO-4-PHENYLBUTANE-1-DIAZONIUM, pyrogenic exotoxin B | | Authors: | Ziomek, E, Sivaraman, J, Doran, J, Menard, R, Cygler, M. | | Deposit date: | 2003-06-27 | | Release date: | 2004-09-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of autoprocessing of the streptococcal pyrogenic exotoxin B (speB). Crystal structure of the proenzyme-inhibitor complex
To be published
|
|
1Q18
 
 | | Crystal structure of E.coli glucokinase (Glk) | | Descriptor: | Glucokinase | | Authors: | Lunin, V.V, Li, Y, Schrag, J.D, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-07-18 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose.
J.Bacteriol., 186, 2004
|
|
1HN0
 
 | |
1HBT
 
 | |
1HMU
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1F2E
 
 | | STRUCTURE OF SPHINGOMONAD, GLUTATHIONE S-TRANSFERASE COMPLEXED WITH GLUTATHIONE | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Nishio, T, Watanabe, T, Patel, A, Wang, Y, Lau, P.C.K, Grochulski, P, Li, Y, Cygler, M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-06-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Properties of a Sphingomonad and Marine Bacterium Beta-Class Glutathione S-Transferases and Crystal Structure of the Former Complex with Glutathione
To be published
|
|
1HMW
 
 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-05 | | Release date: | 2001-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1SBZ
 
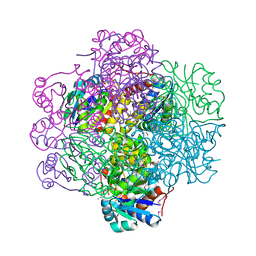 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
1HM2
 
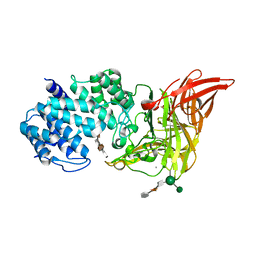 | | ACTIVE SITE OF CHONDROITINASE AC LYASE REVEALED BY THE STRUCTURE OF ENZYME-OLIGOSACCHARIDE COMPLEXES AND MUTAGENESIS | | Descriptor: | 2-O-methyl-beta-L-fucopyranose-(1-4)-beta-D-xylopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-2)-[alpha-L-rhamnopyranose-(1-4)]alpha-D-mannopyranose, CALCIUM ION, CHONDROITINASE AC, ... | | Authors: | Huang, W, Boju, L, Tkalec, L, Su, H, Yang, H.O, Gunay, N.S, Linhardt, R.J, Kim, Y.S, Matte, A, Cygler, M. | | Deposit date: | 2000-12-04 | | Release date: | 2001-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis.
Biochemistry, 40, 2001
|
|
1EOL
 
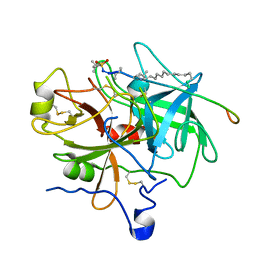 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P628 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
1FG7
 
 | |
1FC5
 
 | |
1PS6
 
 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-HYDROXY-L-THREONINE-5-MONOPHOSPHATE, 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1FC4
 
 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1PRZ
 
 | | Crystal structure of pseudouridine synthase RluD catalytic module | | Descriptor: | Ribosomal large subunit pseudouridine synthase D | | Authors: | Sivaraman, J, Iannuzzi, P, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the RluD pseudouridine Synthase catalytic module, an
enzyme that modifies 23S rRNA and is essential for normal cell growth of Escherichia coli
J.Mol.Biol., 335, 2003
|
|
