7A3T
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | Core protein, EDE1 C8 antibody Fab fragment, GLYCEROL, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3U
 
 | | Crystal structure of Zika virus envelope glycoprotein in complex with the divalent F(ab')2 fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | EDE1 C10 antibody divalent F(ab')2 fragment, EDE1 C10 divalent F(ab')2 fragment, Envelope protein | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3P
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Single Chain Variable Fragment | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3S
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, SULFATE ION | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Navarro-Sanchez, E, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3Q
 
 | | Crystal structure of dengue 4 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
1HAF
 
 | |
1HAE
 
 | |
1HHK
 
 | | THE ANTIGENIC IDENTITY OF PEPTIDE(SLASH)MHC COMPLEXES: A COMPARISON OF THE CONFORMATION OF FIVE PEPTIDES PRESENTED BY HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN (RESIDUES 11-19) | | Authors: | Madden, D.R, Garboczi, D.N, Wiley, D.C. | | Deposit date: | 1993-06-30 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2.
Cell(Cambridge,Mass.), 75, 1993
|
|
1KB5
 
 | | MURINE T-CELL RECEPTOR VARIABLE DOMAIN/FAB COMPLEX | | Descriptor: | ANTIBODY DESIRE-1, KB5-C20 T-CELL ANTIGEN RECEPTOR | | Authors: | Housset, D, Mazza, G, Gregoire, C, Piras, C, Malissen, B, Fontecilla-Camps, J.C. | | Deposit date: | 1997-04-06 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain.
EMBO J., 16, 1997
|
|
1KES
 
 | | CONFORMATION OF KERATAN SULPHATE | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose-(1-3)-6-O-sulfo-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose-(1-3)-6-O-sulfo-beta-D-galactopyranose | | Authors: | Arnott, S. | | Deposit date: | 1978-05-23 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-07 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Conformation of keratan sulphate.
J.Mol.Biol., 88, 1974
|
|
1ITH
 
 | | STRUCTURE DETERMINATION AND REFINEMENT OF HOMOTETRAMERIC HEMOGLOBIN FROM URECHIS CAUPO AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | CYANIDE ION, HEMOGLOBIN (CYANO MET), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hackert, M, Kolatkar, P, Ernst, S.R, Ogata, C.M, Hendrickson, W.A, Merritt, E.A, Phizackerley, R.P. | | Deposit date: | 1991-12-03 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure determination and refinement of homotetrameric hemoglobin from Urechis caupo at 2.5 A resolution.
Acta Crystallogr.,Sect.B, 48, 1992
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
1MYT
 
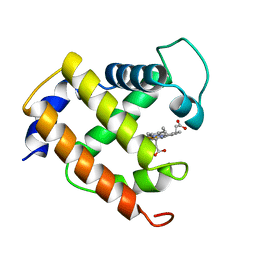 | | CRYSTAL STRUCTURE TO 1.74 ANGSTROMS RESOLUTION OF METMYOGLOBIN FROM YELLOWFIN TUNA (THUNNUS ALBACARES): AN EXAMPLE OF A MYOGLOBIN LACKING THE D HELIX | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Birnbaum, G.I, Evans, S.V, Przybylska, M, Rose, D.R. | | Deposit date: | 1991-05-06 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | 1.70 A resolution structure of myoglobin from yellowfin tuna. An example of a myoglobin lacking the D helix.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1DPI
 
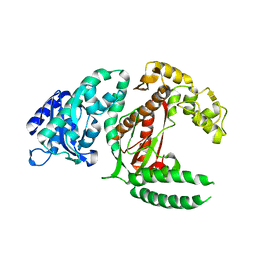 | |
3SGA
 
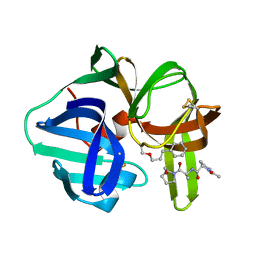 | |
2F3Y
 
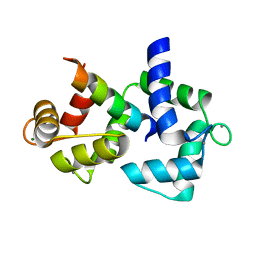 | | Calmodulin/IQ domain complex | | Descriptor: | CALCIUM ION, Calmodulin, MAGNESIUM ION, ... | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
2F3Z
 
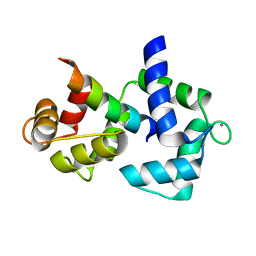 | | Calmodulin/IQ-AA domain complex | | Descriptor: | CALCIUM ION, Calmodulin, Voltage-dependent L-type calcium channel alpha-1C subunit | | Authors: | Fallon, J.L, Quiocho, F.A. | | Deposit date: | 2005-11-22 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Calmodulin Bound to the Hydrophobic IQ Domain of the Cardiac Ca(v)1.2 Calcium Channel.
Structure, 13, 2005
|
|
2GBP
 
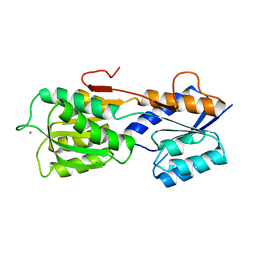 | |
1VHN
 
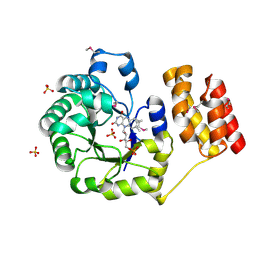 | |
1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
1VIO
 
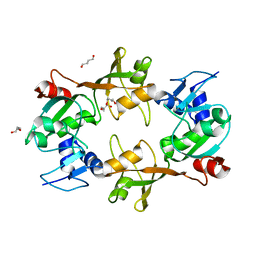 | | Crystal structure of pseudouridylate synthase | | Descriptor: | 1,4-BUTANEDIOL, Ribosomal small subunit pseudouridine synthase A | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1VRK
 
 | |
8BFU
 
 | | Crystal structure of the apo p110alpha catalytic subunit from homo sapiens | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2022-10-26 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
6FMF
 
 | | Neuropilin-1 b1 domain in complex with EG01377; 2.8 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1, trifluoroacetic acid | | Authors: | Yelland, T, Djordjevic, S, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
6FMC
 
 | | Neuropilin1-b1 domain in complex with EG01377, 0.9 Angstrom structure | | Descriptor: | (2~{S})-2-[[3-[[5-[4-(aminomethyl)phenyl]-1-benzofuran-7-yl]sulfonylamino]thiophen-2-yl]carbonylamino]-5-carbamimidamido-pentanoic acid, Neuropilin-1 | | Authors: | Yelland, T, Djordjevic, S, Fotinou, K, Selwood, D, Zachary, I, Frankel, P. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Small Molecule Neuropilin-1 Antagonists Combine Antiangiogenic and Antitumor Activity with Immune Modulation through Reduction of Transforming Growth Factor Beta (TGF beta ) Production in Regulatory T-Cells.
J. Med. Chem., 61, 2018
|
|
