3MFN
 
 | | Dfer_2879 protein of unknown function from Dyadobacter fermentans | | Descriptor: | ACETATE ION, Uncharacterized protein | | Authors: | Osipiuk, J, Xu, X, Cui, H, Chin, S, Eisen, J, Wu, D, Kerfeld, C, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-02 | | Release date: | 2010-04-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | X-ray crystal structure of Dfer_2879 protein of unknown function from Dyadobacter fermentans.
To be Published
|
|
3LLV
 
 | | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A | | Descriptor: | Exopolyphosphatase-related protein, PHOSPHATE ION | | Authors: | Stein, A.J, Chang, C, Weger, A, Hendricks, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the NAD(P)-binding domain of an Exopolyphosphatase-related protein from Archaeoglobus fulgidus to 1.7A
To be Published
|
|
3GJZ
 
 | | Crystal structure of microcin immunity protein MccF from Bacillus anthracis str. Ames | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Kwon, K, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-09 | | Release date: | 2009-04-14 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
3PFM
 
 | | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf | | Descriptor: | GGDEF domain protein | | Authors: | Nocek, B, Stein, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf
TO BE PUBLISHED
|
|
3PU9
 
 | | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein serine/threonine phosphatase | | Authors: | Nocek, B, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745
TO BE PUBLISHED
|
|
3PYZ
 
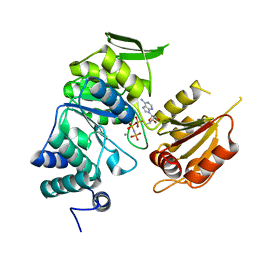 | | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase complexed with AMPPNP and Mn ion from Yersinia pestis c092 | | Descriptor: | Bifunctional folylpolyglutamate synthase/dihydrofolate synthase, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nocek, B, Makowska-Grzyska, M, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bifunctional folylpolyglutamate synthase/dihydrofolate synthase complexed with AMPpnp and Mn ion from Yersinia pestis c092
TO BE PUBLISHED
|
|
3PGY
 
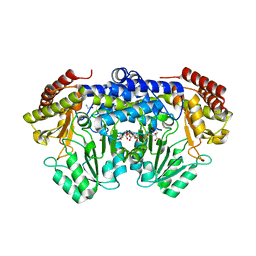 | | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCINE, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant
To be Published
|
|
3PNN
 
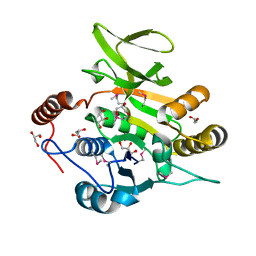 | |
3PNS
 
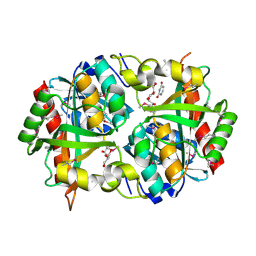 | | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3QOM
 
 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum | | Descriptor: | 6-phospho-beta-glucosidase, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Michalska, K, Hatzos-Skintges, C, Bearden, J, Kohler, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QOK
 
 | |
3QTT
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine | | Descriptor: | BETA-ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-23 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine.
To be Published
|
|
3QN3
 
 | | Phosphopyruvate hydratase from Campylobacter jejuni. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Enolase, GLYCEROL, ... | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Phosphopyruvate hydratase from Campylobacter jejuni.
To be Published
|
|
3Q62
 
 | | Crystal Structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase | | Authors: | Zhang, R, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from
Yersinia pestis
To be Published
|
|
3QBO
 
 | | Crystal structure of phosphoserine aminotransferase from Yersinia pestis CO92 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase | | Authors: | Nocek, B, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-13 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of phosphoserine aminotransferase from Yersinia pestis CO92
TO BE PUBLISHED
|
|
3Q7I
 
 | | Glucose-6-phosphate isomerase from Francisella tularensis complexed with 6-phosphogluconic acid. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-PHOSPHOGLUCONIC ACID, CALCIUM ION, ... | | Authors: | Osipiuk, J, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-01-04 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Glucose-6-phosphate isomerase from Francisella tularensis.
To be Published
|
|
3QU1
 
 | | Peptide deformylase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Peptide deformylase 2, SULFATE ION, ... | | Authors: | Osipiuk, J, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide deformylase from Vibrio cholerae.
To be Published
|
|
3QWU
 
 | | Putative ATP-dependent DNA ligase from Aquifex aeolicus. | | Descriptor: | ADENOSINE, CALCIUM ION, DNA ligase, ... | | Authors: | Osipiuk, J, Quartey, P, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-28 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Putative ATP-dependent DNA ligase from Aquifex aeolicus.
To be Published
|
|
3QOO
 
 | | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans
TO BE PUBLISHED
|
|
3RPC
 
 | |
3RQ5
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with CoA | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, COENZYME A, GLYCEROL | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQH
 
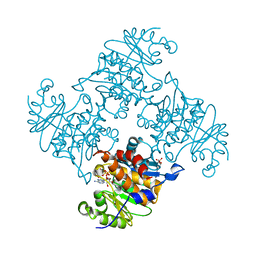 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P6-Di(adenosine-5') hexaphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, MAGNESIUM ION, P1,P6-Di(adenosine-5') hexaphosphate | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RRI
 
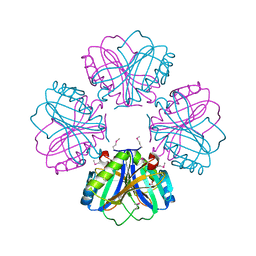 | |
3R6D
 
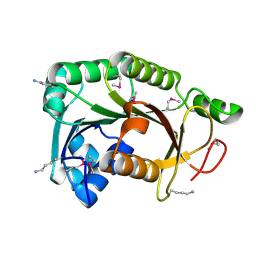 | |
3R7A
 
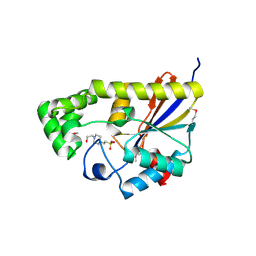 | | Crystal structure of phosphoglycerate mutase from Bacillus anthracis str. Sterne | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phosphoglycerate mutase, putative | | Authors: | Chang, C, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-22 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Crystal structure of phosphoglycerate mutase from Bacillus anthracis str. Sterne
To be Published
|
|
