7C1K
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
5UAN
 
 | | Crystal structure of multi-domain RAR-beta-RXR-alpha heterodimer on DNA | | Descriptor: | (9cis)-retinoic acid, DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*G)-3'), ... | | Authors: | Chandra, V, Wu, D, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-12-19 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | The quaternary architecture of RAR beta-RXR alpha heterodimer facilitates domain-domain signal transmission.
Nat Commun, 8, 2017
|
|
4N0A
 
 | | Crystal structure of Lsm2-3-Pat1C complex from Saccharomyces cerevisiae | | Descriptor: | DNA topoisomerase 2-associated protein PAT1, U6 snRNA-associated Sm-like protein LSm2, U6 snRNA-associated Sm-like protein LSm3 | | Authors: | Wu, D.H. | | Deposit date: | 2013-10-01 | | Release date: | 2013-12-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation
Cell Res., 24, 2014
|
|
8YJ3
 
 | | N17.1.2 recognition of NRAS neoantigens | | Descriptor: | tcr alpha, tcr beta | | Authors: | Wu, D.C, Mariuzza, R.A. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Sci Adv, 10, 2024
|
|
8EY5
 
 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 3 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8YJ2
 
 | | N17.1.2 recognition of NRAS neoantigens | | Descriptor: | Beta-2-microglobulin, ILE-LEU-ASP-THR-ALA-GLY-ARG-GLU-GLU-TYR, human leukocyte antigen, ... | | Authors: | Wu, D.C, Mariuzza, R.A. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Sci Adv, 10, 2024
|
|
8YIV
 
 | | N17.1.2 recognition of NRAS neoantigens | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Wu, D.C, Mariuzza, R.A. | | Deposit date: | 2024-02-29 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structural characterization and AlphaFold modeling of human T cell receptor recognition of NRAS cancer neoantigens.
Sci Adv, 10, 2024
|
|
8EW4
 
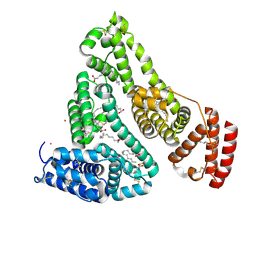 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 1 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Unciano, J, Lea, K, Kim, L, Lenkiewicz, J, Starban, I, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
8EW7
 
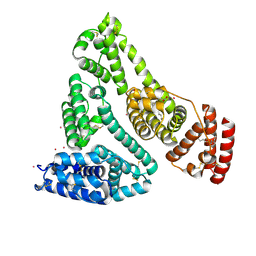 | | Human Serum Albumin with Cobalt (II) and Myristic Acid - crystal 2 | | Descriptor: | COBALT (II) ION, MYRISTIC ACID, Serum albumin | | Authors: | Gucwa, M, Cooper, D.R, Stewart, A.J, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and biochemical characterisation of Co2+-binding sites on serum albumins and their interplay with fatty acids
Chem Sci, 14, 2023
|
|
7MBL
 
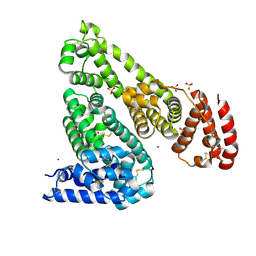 | | Crystal structure of Equine Serum Albumin in complex with Cobalt (II) | | Descriptor: | COBALT (II) ION, SULFATE ION, Serum albumin | | Authors: | Shabalin, I.G, Czub, M.P, Handing, K.B, Cymborowski, M.T, Grabowski, M, Cooper, D.R, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterisation of Co 2+ -binding sites on serum albumins and their interplay with fatty acids.
Chem Sci, 14, 2023
|
|
7YLA
 
 | | Cryo-EM structure of 50S-HflX complex | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Damu, W, Ning, G. | | Deposit date: | 2022-07-25 | | Release date: | 2023-01-04 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM Structure of the 50S-HflX Complex Reveals a Novel Mechanism of Antibiotic Resistance in E. coli
Biorxiv, 2022
|
|
7QVJ
 
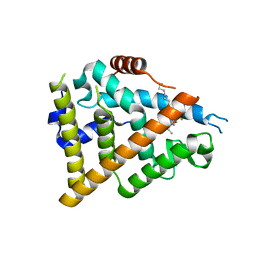 | | ESTROGEN RECEPTOR ALPHA IN COMPLEX WITH COMPOUND 29 | | Descriptor: | 2,2-bis(fluoranyl)-3-[(1~{R},3~{R})-1-[6-fluoranyl-3-[2-(3-fluoranylpropylamino)ethoxy]-2-methyl-phenyl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]propan-1-ol, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
7QVL
 
 | | OESTROGEN RECEPTOR LIGAND BINDING DOMAIN IN COMPLEX WITH COMPOUND 38 | | Descriptor: | (2~{R})-3-[(1~{R},3~{R})-1-[5-fluoranyl-2-[2-(3-fluoranylpropylamino)ethoxy]-3-methyl-pyridin-4-yl]-3-methyl-1,3,4,9-tetrahydropyrido[3,4-b]indol-2-yl]-2-methyl-propanoic acid, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Potent and Orally Bioavailable Zwitterionic Series of Selective Estrogen Receptor Degrader-Antagonists.
J.Med.Chem., 66, 2023
|
|
6Z45
 
 | | CDK9-Cyclin-T1 complex bound by compound 24 | | Descriptor: | (1~{S},3~{R})-3-acetamido-~{N}-[5-chloranyl-4-(5,5-dimethyl-4,6-dihydropyrrolo[1,2-b]pyrazol-3-yl)pyridin-2-yl]cyclohexane-1-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cyclin-T1, ... | | Authors: | Ferguson, A, Collie, G.W. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Discovery of AZD4573, a Potent and Selective Inhibitor of CDK9 That Enables Short Duration of Target Engagement for the Treatment of Hematological Malignancies.
J.Med.Chem., 63, 2020
|
|
9QT6
 
 | | Crystal structure of HPK1 T165E/S171E in complex with pyrazine carboxamide inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[(1,3-dimethylpyrazol-4-yl)amino]-5-(methylamino)-6-(3-methylimidazo[4,5-c]pyridin-7-yl)pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Schimpl, M. | | Deposit date: | 2025-04-08 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
9J0K
 
 | | An agonist(compound 14e)of Thyroid Hormone Receptor B | | Descriptor: | 2-[[7-[3-(aminomethyl)phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B, Li, Y. | | Deposit date: | 2024-08-02 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Discovery of a Potent, Selective, and Multiple His435 Mutation-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 2025
|
|
9IX5
 
 | | An agonist(compound 15n) of Thyroid Hormone Receptor B | | Descriptor: | 2-[(1-methoxy-7-naphthalen-2-yloxy-4-oxidanyl-isoquinolin-3-yl)carbonylamino]ethanoic acid, Nuclear receptor coactivator 2, Thyroid hormone receptor beta | | Authors: | Yao, B, Li, Y. | | Deposit date: | 2024-07-26 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a Potent, Selective, and Multiple His435 Mutation-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 2025
|
|
8HIP
 
 | | dsRNA transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Systemic RNA interference defective protein 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HKE
 
 | | dsRNA transporter | | Descriptor: | RNA (37-MER), Systemic RNA interference defective protein 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-11-29 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7FCP
 
 | |
7FCQ
 
 | |
5Y93
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[5-[(5-bromanyl-2-methoxy-phenyl)sulfonylamino]-3-methyl-1,2-benzoxazol-6-yl]oxy]-N-(2-morpholin-4-ylethyl)ethanamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Song, M, Wang, C. | | Deposit date: | 2017-08-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Discovery and Optimization of Benzo[ d]isoxazole Derivatives as Potent and Selective BET Inhibitors for Potential Treatment of Castration-Resistant Prostate Cancer (CRPC)
J. Med. Chem., 61, 2018
|
|
8HSB
 
 | |
9ODR
 
 | | Structure of CRBN TBD bound to compound C1 | | Descriptor: | (3S)-3-(6-oxo-6,8-dihydro-2H,7H-spiro[furo[2,3-e]isoindole-3,4'-piperidin]-7-yl)piperidine-2,6-dione, Protein cereblon, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2025-04-27 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
9ODS
 
 | | Structure of CRBN TBD bound to compound C3 | | Descriptor: | (1P)-1-[(4S)-8H-spiro[furo[2,3-c]imidazo[1,2-a]pyridine-7,4'-piperidin]-3-yl]pyrimidine-2,4(1H,3H)-dione, Protein cereblon, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2025-04-27 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
