6Z0C
 
 | | Structure of in silico modelled artificial Maquette-3 protein | | Descriptor: | Maquette-3, POTASSIUM ION | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M.E, Asami, S, Mader, S.L, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of buried charged networks in artificial proteins
Nat Commun, 12, 2021
|
|
9GLF
 
 | | Anthraquinone Pigment Production Regulated by Cinnamic Acid | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AntI, ... | | Authors: | Su, L, Schmalhofer, M, Grammbitter, G.L.C, Paczia, N, Glatter, T, Groll, M, Bode, H.B. | | Deposit date: | 2024-08-27 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Isopropylstilbene Precursor Cinnamic Acid Inhibits Anthraquinone Pigment Production by Targeting AntI.
J.Am.Chem.Soc., 147, 2025
|
|
5T71
 
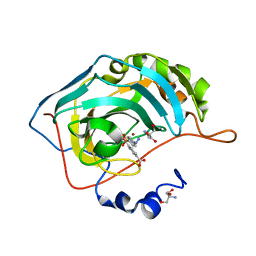 | | Human carboanhydrase F131C_C206S double mutant in complex with SA-2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-diazenyl]benzene-1-sulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5T75
 
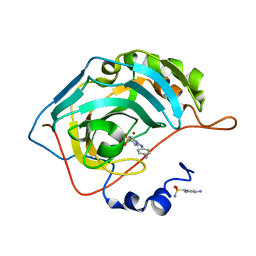 | | Human carbonic anhydrase II G132C_C206S double mutant in complex with SA-2 | | Descriptor: | 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
5T74
 
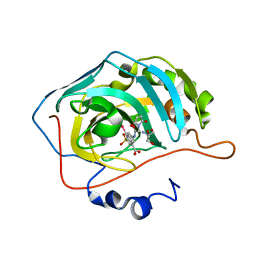 | | Human carboanhydrase F131C_C206S double mutant in complex with 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[2,5-bis(oxidanylidene)pyrrol-1-yl]-~{N}-(4-sulfamoylphenyl)ethanamide, 4-(HYDROXYMERCURY)BENZOIC ACID, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
6EK4
 
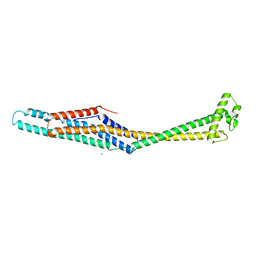 | | PaxB from Photorhabdus luminescens | | Descriptor: | PaxB, SODIUM ION | | Authors: | Braeuning, B, Groll, M. | | Deposit date: | 2017-09-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
6EL1
 
 | | YaxAB pore complex | | Descriptor: | YaxA, YaxB | | Authors: | Braeuning, B, Bertosin, E, Dietz, H, Groll, M. | | Deposit date: | 2017-09-27 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
9EY9
 
 | | Yeast 20S proteasome in complex with a sybactin derivative (PheSyr) | | Descriptor: | MAGNESIUM ION, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Praeve, L, Kuttenlochner, W, Tabak, W.W.A, Langer, C, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2024-04-09 | | Release date: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bioengineering of syrbactin megasynthetases for immunoproteasome inhibitor production
Chem, 10, 2024
|
|
6EK7
 
 | | YaxA from Yersinia enterocolitica | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, YaxA | | Authors: | Braeuning, B, Groll, M. | | Deposit date: | 2017-09-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB.
Nat Commun, 9, 2018
|
|
5T72
 
 | | Human carboanhydrase F131C_C206S double mutant in complex with 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(HYDROXYMERCURY)BENZOIC ACID, 4-[(E)-(4-aminophenyl)diazenyl]benzenesulfonamide, ... | | Authors: | DuBay, K.H, Iwan, K, Osorio-Planes, L, Geissler, P, Groll, M, Trauner, D, Broichhagen, J. | | Deposit date: | 2016-09-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Predictive Approach for the Optical Control of Carbonic Anhydrase II Activity.
ACS Chem. Biol., 13, 2018
|
|
8ANP
 
 | | Legionella effector Lem3 mutant D190A in complex with Mg2+ | | Descriptor: | MAGNESIUM ION, Phosphocholine hydrolase Lem3, SULFATE ION, ... | | Authors: | Kaspers, M.S, Pogenberg, V, Ernst, S, Ecker, F, Pett, C, Ochtrop, P, Hedberg, C, Groll, M, Itzen, A. | | Deposit date: | 2022-08-05 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dephosphocholination by Legionella effector Lem3 functions through remodelling of the switch II region of Rab1b.
Nat Commun, 14, 2023
|
|
8A7K
 
 | | PcIDS1 in complex with Mg2+/Mn2+, IPP, and ZOL | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7A
 
 | | PcIDS1 in complex with Mg2+ and 3-Br-GPP | | Descriptor: | Isoprenyl diphosphate synthase, MAGNESIUM ION, [(2Z)-3-bromanyl-7-methyl-octa-2,6-dienyl] phosphono hydrogen phosphate | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7L
 
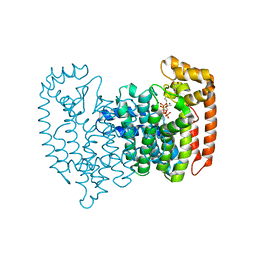 | | PcIDS1 in complex with Mg2+, GPP, and ZOL | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7C
 
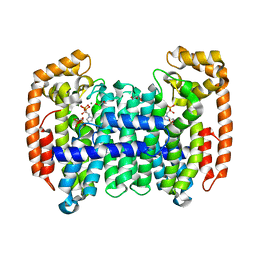 | | PcIDS1 in complex with Mg2+, IPP, and ZOL | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7J
 
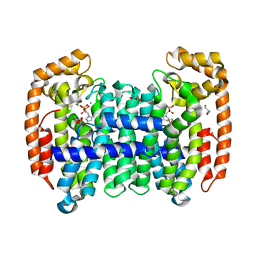 | | PcIDS1 in complex with Mn2+, IPP, and ZOL | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7R
 
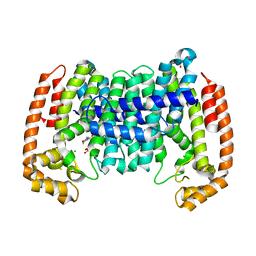 | | PcIDS1_I420A in complex with Mg2+ and GPP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GERANYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7U
 
 | | PcIDS1_F315D in complex with Mg2+ and GPP | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, F, Groll, M. | | Deposit date: | 2022-06-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A7B
 
 | | PcIDS1_D319N in complex with Mg2+ and 3-Br-GPP | | Descriptor: | CARBONATE ION, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8A78
 
 | | PcIDS1_F315A in complex with Mg2+/Mn2+ and GPP | | Descriptor: | GERANYL DIPHOSPHATE, GLYCEROL, Isoprenyl diphosphate synthase, ... | | Authors: | Ecker, F, Boland, W, Groll, M. | | Deposit date: | 2022-06-20 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-dependent enzyme symmetry guides the biosynthetic flux of terpene precursors.
Nat.Chem., 15, 2023
|
|
8RYD
 
 | | AzeJ in complex with SAH from Pseudomonas aeruginosa | | Descriptor: | Class I SAM-dependent methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Klaubert, T.J, Gellner, J, Bernard, C, Effert, J, Lombard, C, Kaila, V.R.I, Bode, H.B, Li, Y, Groll, M. | | Deposit date: | 2024-02-08 | | Release date: | 2025-01-01 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for azetidine-2-carboxylic acid biosynthesis.
Nat Commun, 16, 2025
|
|
7O2L
 
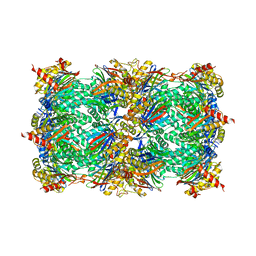 | | Yeast 20S proteasome in complex with the covalently bound inhibitor b-lactone (2R,3S)-3-isopropyl-4-oxo-2-oxetane-carboxylate (IOC) | | Descriptor: | (2 {R},3 {S})-3-methanoyl-4-methyl-2-hydroxy-pentanoic acid, 20S proteasome, BJ4_G0020160.mRNA.1.CDS.1, ... | | Authors: | Shi, Y.M, Hirschmann, M, Shi, Y.N, Shabbir, A, Abebew, D, Tobias, N.J, Gruen, P, Crames, J.J, Poeschel, L, Kuttenlochner, W, Richter, C, Herrmann, J, Mueller, R, Thanwisai, A, Pidot, S.J, Stinear, T.P, Groll, M, Kim, Y, Bode, H. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria.
Nat.Chem., 14, 2022
|
|
6ZOU
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB333 | | Descriptor: | 11-methyl-~{N}-[(2~{S},3~{R})-1-[[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]amino]-3-oxidanyl-1-oxidanylidene-butan-2-yl]dodecanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-07 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6ZP8
 
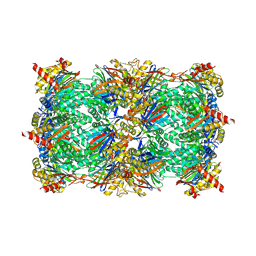 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB335 | | Descriptor: | (2~{S},3~{R})-~{N}-[(5~{S},8~{S},10~{S})-5-methyl-10-oxidanyl-2,7-bis(oxidanylidene)-1,6-diazacyclododec-8-yl]-3-oxidanyl-2-(3-phenylpropanoylamino)butanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
6ZP6
 
 | | Yeast 20S proteasome in complex with glidobactin-like natural product HB334 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Zhao, L, Le Chapelain, C, Brachmann, A.O, Kaiser, M, Groll, M, Bode, H.B. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation, Structure, Biosynthesis and Bioactivity of Glidobactin-like Proteasome Inhibitors from Photorhabdus laumondii.
Chembiochem, 22, 2021
|
|
