2E61
 
 | | Solution structure of the zf-CW domain in zinc finger CW-type PWWP domain protein 1 | | Descriptor: | ZINC ION, Zinc finger CW-type PWWP domain protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-25 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the zinc finger CW domain as a histone modification reader
Structure, 18, 2010
|
|
2CU9
 
 | |
6SP7
 
 | | Crystal Structure of the VIM-2 Acquired Metallo-beta-Lactamase in Complex with Taniborbactam (VNRX-5133) | | Descriptor: | (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, ACETATE ION, Metallo-beta-lactamase VIM-2, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections.
J.Med.Chem., 63, 2020
|
|
6SP6
 
 | | Ultra-high Resolution Crystal Structure of the CTX-M-15 Extended-Spectrum beta-Lactamase in Complex with Taniborbactam (VNRX-5133) | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of Taniborbactam (VNRX-5133): A Broad-Spectrum Serine- and Metallo-beta-lactamase Inhibitor for Carbapenem-Resistant Bacterial Infections.
J.Med.Chem., 63, 2020
|
|
9AZY
 
 | | Crystal structure of PDC-88 beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2024-03-11 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of taniborbactam inhibition of the cefepime-hydrolyzing, partial R2-loop deletion Pseudomonas -derived cephalosporinase variant PDC-88.
Antimicrob.Agents Chemother., 69, 2025
|
|
9AZU
 
 | | Crystal structure of PDC-3 beta-lactamase in complex with taniborbactam | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2024-03-11 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure and mechanism of taniborbactam inhibition of the cefepime-hydrolyzing, partial R2-loop deletion Pseudomonas -derived cephalosporinase variant PDC-88.
Antimicrob.Agents Chemother., 69, 2025
|
|
9AZW
 
 | | Crystal structure of PDC-88 beta-lactamase in complex with taniborbactam | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase | | Authors: | Kumar, V, van den Akker, F. | | Deposit date: | 2024-03-11 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and mechanism of taniborbactam inhibition of the cefepime-hydrolyzing, partial R2-loop deletion Pseudomonas -derived cephalosporinase variant PDC-88.
Antimicrob.Agents Chemother., 69, 2025
|
|
2DX8
 
 | |
2DW4
 
 | |
3AKG
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranobiose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKH
 
 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-1,5-L-arabinofuranotriose | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKI
 
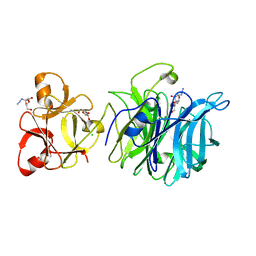 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase complexed with alpha-L-arabinofuranosyl azido | | Descriptor: | (2R,3R,4R,5S)-2-azido-5-(hydroxymethyl)oxolane-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
3AKF
 
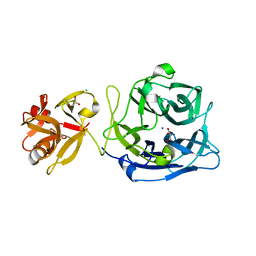 | | Crystal structure of exo-1,5-alpha-L-arabinofuranosidase | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative secreted alpha L-arabinofuranosidase II, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of an Exo-1,5-{alpha}-L-arabinofuranosidase from Streptomyces avermitilis Provides Insights into the Mechanism of Substrate Discrimination between Exo- and Endo-type Enzymes in Glycoside Hydrolase Family 43.
J.Biol.Chem., 285, 2010
|
|
2RS9
 
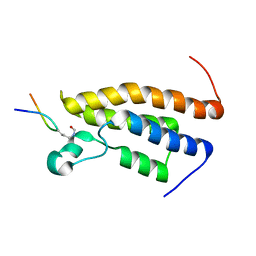 | | Solution structure of the bromodomain of human BRPF1 in complex with histone H4K5ac peptide | | Descriptor: | Acetylated lysine 5 of peptide from Histone H4, Peregrin | | Authors: | Qin, X, Nagashima, T, Umehara, T, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Site-specific histone recognition by the bromodomain of Brpf1 and the role in MOZ/MORF histone acetyltransferase complexes
To be Published
|
|
2DVW
 
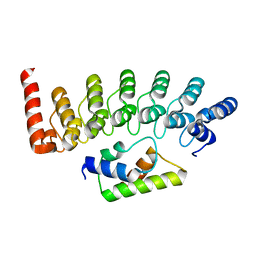 | |
2DWZ
 
 | |
2DZN
 
 | |
2E3K
 
 | |
