5OLP
 
 | | Galacturonidase | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
5OLS
 
 | | Rhamnogalacturonan lyase | | Descriptor: | 4-deoxy-beta-L-threo-hex-4-enopyranuronic acid-(1-2)-alpha-L-rhamnopyranose-(1-4)-beta-D-galactopyranuronic acid, CALCIUM ION, Rhamnogalacturonan lyase, ... | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
3E74
 
 | |
3OCZ
 
 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase Complexed with the inhibitor adenosine 5-O-thiomonophosphate | | Descriptor: | ADENOSINE -5'-THIO-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of the inhibition of class C acid phosphatases by adenosine 5'-phosphorothioate.
Febs J., 278, 2011
|
|
5OPJ
 
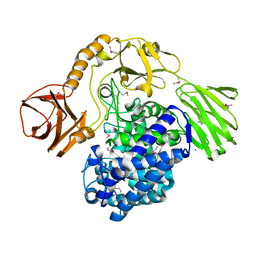 | | Beta-L-arabinofuranosidase | | Descriptor: | Rhamnogalacturonan lyase, ZINC ION, alpha-L-arabinofuranose | | Authors: | Basle, A, Luis, A.S, Gilbert, H.J. | | Deposit date: | 2017-08-10 | | Release date: | 2018-02-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides.
Nat Microbiol, 3, 2018
|
|
4BKM
 
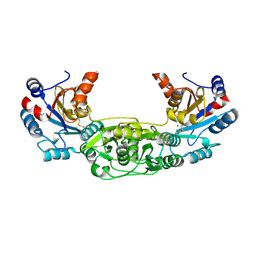 | | Crystal structure of the murine AUM (phosphoglycolate phosphatase) capping domain as a fusion protein with the catalytic core domain of murine chronophin (pyridoxal phosphate phosphatase) | | Descriptor: | MAGNESIUM ION, NITRATE ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Seifried, A, Gohla, A, Schindelin, H. | | Deposit date: | 2013-04-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Evolutionary and Structural Analyses of the Mammalian Haloacid Dehalogenase-Type Phosphatases Aum and Chronophin Provide Insight Into the Basis of Their Different Substrate Specificities.
J.Biol.Chem., 289, 2014
|
|
4F14
 
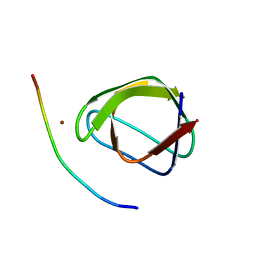 | |
8TU5
 
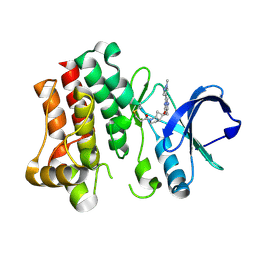 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 27 | | Descriptor: | 1-[(1S,5S,6S)-6-methyl-6-{[(6M,8R)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}-2-azabicyclo[3.2.0]heptan-2-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8TU3
 
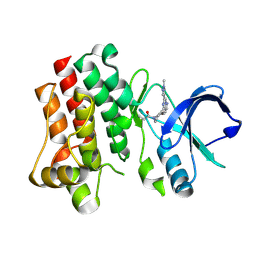 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 10 | | Descriptor: | 1-[(4R)-4-{[(6P,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}azepan-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8TU4
 
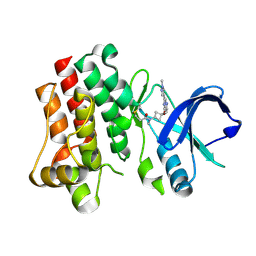 | | Bruton's tyrosine kinase in complex with covalent inhibitor compound 25 | | Descriptor: | N-methyl-N-[(1S,3r)-3-methyl-3-{[(6M,8S)-6-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrazin-4-yl]oxy}cyclobutyl]propanamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M. | | Deposit date: | 2023-08-15 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Preclinical Characterization of BIIB129, a Covalent, Selective, and Brain-Penetrant BTK Inhibitor for the Treatment of Multiple Sclerosis.
J.Med.Chem., 67, 2024
|
|
8IU1
 
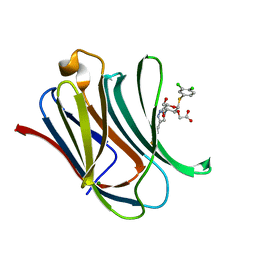 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2R,3R,4S,5R,6R)-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-5-oxidanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-yl]oxyethanoic acid, Galectin-3, MAGNESIUM ION | | Authors: | Jinal, S, Amit, K, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
8ITZ
 
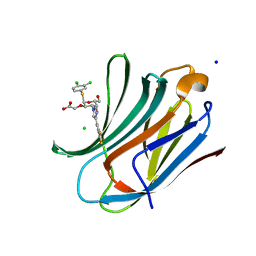 | | Crystal structure of human Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2R,3R,4S,5R,6R)-2-(3,4-dichlorophenyl)sulfanyl-6-(hydroxymethyl)-5-oxidanyl-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-3-yl]oxyethanoic acid, CHLORIDE ION, Galectin-3, ... | | Authors: | Jinal, S, Amit, K, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
8ITX
 
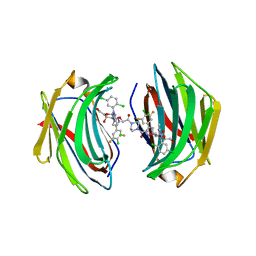 | | Crystal structure of human Galectin-3 in complex with small molecule inhibitor | | Descriptor: | 2-[(2S,3R,4S,5R,6R)-2-[2-[2,5-bis(chloranyl)phenyl]-5-methyl-1,2,4-triazol-3-yl]-4-[4-[4-chloranyl-3,5-bis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-6-(hydroxymethyl)-5-oxidanyl-oxan-3-yl]oxyethanoic acid, CHLORIDE ION, Galectin-3, ... | | Authors: | Jinal, S, Ghosh, K. | | Deposit date: | 2023-03-23 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Discovery and Exploration of Monosaccharide Linked Dimers to Target Fibrosis
To Be Published
|
|
6T3J
 
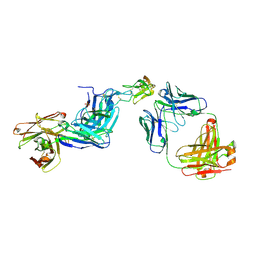 | | Dual Epitope Targeting by Anti-DR5 Antibodies | | Descriptor: | IgG1-hDR5-01-Heavy Chain, IgG1-hDR5-01-Light Chain, IgG1-hDR5-05-Heavy Chain, ... | | Authors: | Tauchert, M.J, Augustin, M, Krapp, S, Overdijk, M.B, Breij, E.C.W, Hibbert, R.G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Dual Epitope Targeting and Enhanced Hexamerization by DR5 Antibodies as a Novel Approach to Induce Potent Antitumor Activity Through DR5 Agonism.
Mol.Cancer Ther., 19, 2020
|
|
1CUR
 
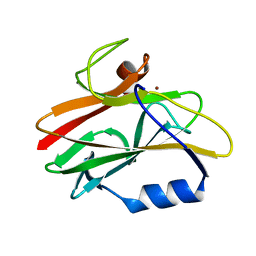 | | REDUCED RUSTICYANIN, NMR | | Descriptor: | COPPER (II) ION, CU(I) RUSTICYANIN | | Authors: | Botuyan, M.V, Dyson, H.J. | | Deposit date: | 1996-04-19 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cu(I) rusticyanin from Thiobacillus ferrooxidans: structural basis for the extreme acid stability and redox potential.
J.Mol.Biol., 263, 1996
|
|
6RK0
 
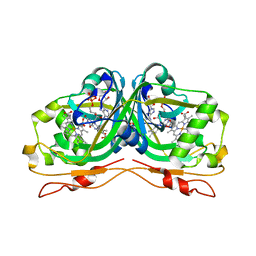 | | Structure of the Flavocytochrome Anf3 from Azotobacter vinelandii | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Murray, J.W, Varghese, F, Kabasakal, B. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A low-potential terminal oxidase associated with the iron-only nitrogenase from the nitrogen-fixing bacteriumAzotobacter vinelandii.
J.Biol.Chem., 294, 2019
|
|
5FU4
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOS
 
 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
6FPG
 
 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
5FU3
 
 | | The complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition | | Descriptor: | CBM74-RFGH5, SODIUM ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Basle, A, Luis, A.S, Venditto, I, Gilbert, H.J. | | Deposit date: | 2016-01-20 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6HE2
 
 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HDX
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with R3-HIB-AMP | | Descriptor: | (2R)-3-HYDROXY-2-METHYLPROPANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] (2~{R})-2-methyl-3-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6HE0
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA in the thioesterfication state | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
6H3P
 
 | |
