4YIR
 
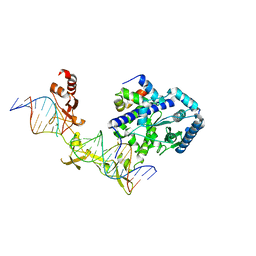 | | Crystal structure of Rad4-Rad23 crosslinked to an undamaged DNA | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*AP*GP*CP*G*GP*GP*GP*AP*TP*GP*TP*CP*GP*AP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*(G47)P*AP*CP*AP*TP*CP*CP*CP*CP*CP*GP*CP*TP*AP*CP*AP*A)-3'), DNA repair protein RAD4, ... | | Authors: | Min, J.-H, Chen, X, Kim, Y. | | Deposit date: | 2015-03-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.0501 Å) | | Cite: | Kinetic gating mechanism of DNA damage recognition by Rad4/XPC.
Nat Commun, 6, 2015
|
|
358D
 
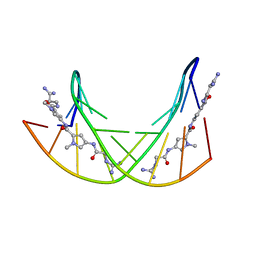 | |
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | Authors: | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | Deposit date: | 1998-05-27 | | Release date: | 1998-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
4TY9
 
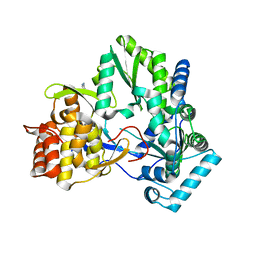 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 5-(trifluoromethyl)pyridin-2-amine, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYB
 
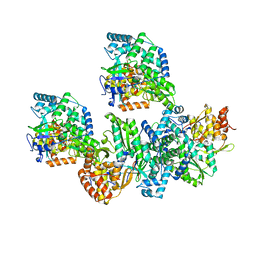 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | (2R)-morpholin-4-yl(phenyl)ethanenitrile, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
4TYA
 
 | | An Ligand-observed Mass Spectrometry-based Approach Integrated into the Fragment Based Lead Discovery Pipeline | | Descriptor: | 4-(trifluoromethyl)benzoic acid, Polyprotein | | Authors: | Shui, W, Yang, C, Lin, J, Chen, X, Qin, S, Chen, S. | | Deposit date: | 2014-07-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A ligand-observed mass spectrometry approach integrated into the fragment based lead discovery pipeline
Sci Rep, 5, 2015
|
|
2Z59
 
 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
7SUD
 
 | | CryoEM structure of DNA-PK complex VIII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-dependent protein kinase catalytic subunit, MAGNESIUM ION, ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
7SU3
 
 | | CryoEM structure of DNA-PK complex VII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), ... | | Authors: | Chen, X, Liu, L, Gellert, M, Yang, W. | | Deposit date: | 2021-11-16 | | Release date: | 2022-01-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Autophosphorylation transforms DNA-PK from protecting to processing DNA ends.
Mol.Cell, 82, 2022
|
|
8YRP
 
 | | SARS-CoV-2 Delta Spike in complex with JM-1A | | Descriptor: | JM-1A Heavy Chain, JM-1A Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YRO
 
 | | SARS-CoV-2 Delta Spike in complex with JL-8C | | Descriptor: | JL-8C Heavy Chain, JL-8C Light Chain, Spike glycoprotein | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2024-03-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
6U19
 
 | |
2N3T
 
 | |
2N3U
 
 | |
2N3W
 
 | |
2N3V
 
 | |
9D0T
 
 | | Core particle assembly intermediate 1 purified from Saccharomyces cerevisiae | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome activator BLM10, Proteasome maturation factor UMP1,Myosin light chain kinase 2, ... | | Authors: | Chen, X, Kaur, M, Roelofs, J, Walters, K.J. | | Deposit date: | 2024-08-07 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure of Blm10:13S proteasome intermediate reveals parallel assembly pathways for the proteasome core particle.
Biorxiv, 2024
|
|
9KAY
 
 | |
4TTQ
 
 | |
5W77
 
 | | Solution structure of the MYC G-quadruplex bound to small molecule DC-34 | | Descriptor: | 4-[(azepan-1-yl)methyl]-5-hydroxy-2-methyl-N-[4-(trifluoromethyl)phenyl]-1-benzofuran-3-carboxamide, DNA (5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*GP*GP*GP*TP*GP*GP*GP*TP*AP*A)-3'), POTASSIUM ION | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2017-06-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex.
Nat Commun, 9, 2018
|
|
7JKS
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2411a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 gp120 core, The heavy chain of antibody 2411a, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
7JKT
 
 | | Crystal structure of vaccine-elicited broadly neutralizing VRC01-class antibody 2413a in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 gp120 core from strain d45-01dG5, ... | | Authors: | Zhou, T, Chen, X, Kwong, P.D, Mascola, J.R. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Vaccination induces maturation in a mouse model of diverse unmutated VRC01-class precursors to HIV-neutralizing antibodies with >50% breadth.
Immunity, 54, 2021
|
|
6CIJ
 
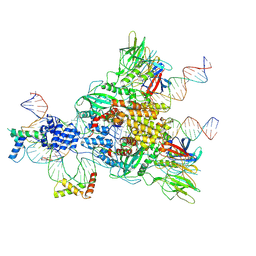 | | Cryo-EM structure of mouse RAG1/2 HFC complex containing partial HMGB1 linker(3.9 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CG0
 
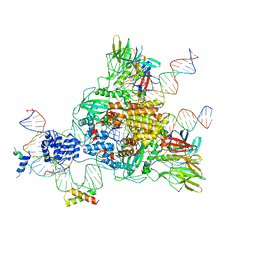 | | Cryo-EM structure of mouse RAG1/2 HFC complex (3.17 A) | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (41-MER), ... | | Authors: | Chen, X, Kim, M, Chuenchor, W, Cui, Y, Zhang, X, Zhou, Z.H, Gellert, M, Yang, W. | | Deposit date: | 2018-02-19 | | Release date: | 2018-04-25 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
8S6I
 
 | | Crystal structure of the human CDKL2 kinase domain with Compound 9 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclin-dependent kinase-like 2, ... | | Authors: | Chen, X, Ni, X, Brooke, L, Bullock, A.N. | | Deposit date: | 2024-02-27 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery and Characterization of a Chemical Probe for Cyclin-Dependent Kinase-Like 2.
Acs Med.Chem.Lett., 15, 2024
|
|
