7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VAH
 
 | |
7WRP
 
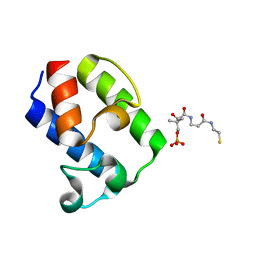 | | Crystal Structure of pks13-ACP domain from Corynebacterium diphtheriae | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase involved in mycolic acid biosynthesis | | Authors: | Liu, X. | | Deposit date: | 2022-01-27 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structures of FadD32 and pks13-ACP domain from Corynebacterium diphtheriae.
Biochem.Biophys.Res.Commun., 590, 2022
|
|
7BV5
 
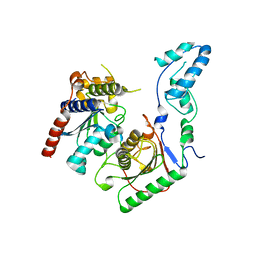 | | Crystal structure of the yeast heterodimeric ADAT2/3 | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase subunit TAD2, tRNA-specific adenosine deaminase subunit TAD3 | | Authors: | Xie, W, Liu, X, Chen, R, Sun, Y, Chen, R, Zhou, J, Tian, Q. | | Deposit date: | 2020-04-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the yeast heterodimeric ADAT2/3 deaminase.
Bmc Biol., 18, 2020
|
|
6IJZ
 
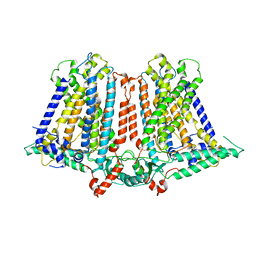 | | Structure of a plant cation channel | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Sun, L, Wang, J, Liu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2.
Nat Commun, 9, 2018
|
|
7EEY
 
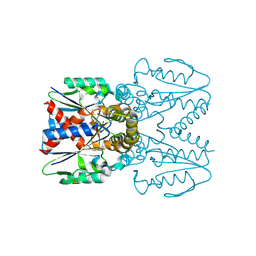 | |
2VAS
 
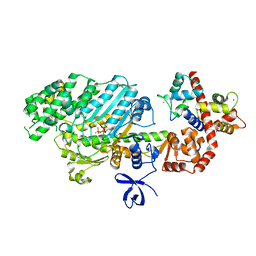 | | Myosin VI (MD-insert2-CaM, Delta-Insert1) Post-rigor state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Menetrey, J, Llinas, P, Cicolari, J, Squires, G, Liu, X, Li, A, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Post-Rigor Structure of Myosin Vi and Implications for the Recovery Stroke.
Embo J., 27, 2008
|
|
3CXS
 
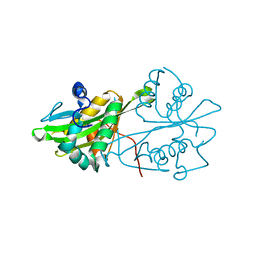 | | Crystal structure of human GNA1 | | Descriptor: | Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
7BU7
 
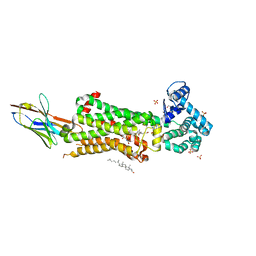 | | Structure of human beta1 adrenergic receptor bound to BI-167107 and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-[(1R)-2-{[1,1-dimethyl-2-(2-methylphenyl)ethyl]amino}-1-hydroxyethyl]-5-hydroxy-2H-1,4-benzoxazin-3(4H)-one, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
7MNW
 
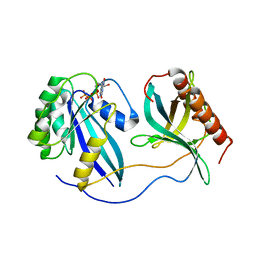 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 1 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MVX
 
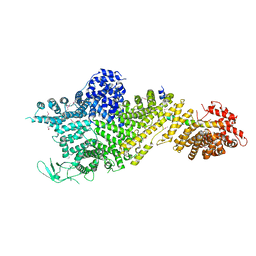 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MNZ
 
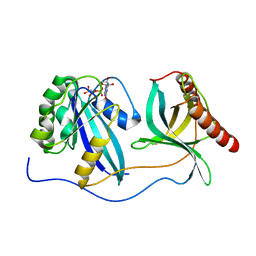 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 4 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNI
 
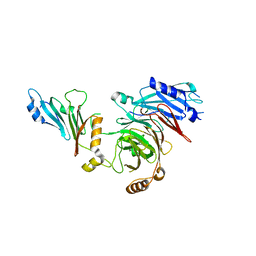 | | Crystal structure of the N-terminal domain of NUP88 in complex with NUP98 C-terminal Autoproteolytic Domain | | Descriptor: | Nuclear pore complex protein Nup88, Nuclear pore complex protein Nup98 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNU
 
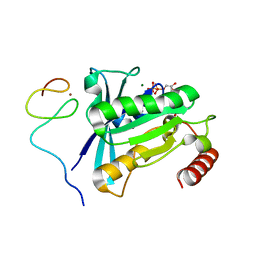 | | Crystal Structure of the ZnF7 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNJ
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO0
 
 | | Crystal Structure of Nucleoporin NUP50 Ran-Binding Domain in Complex with Ran-GPPNHP | | Descriptor: | GTP-binding nuclear protein Ran, MAGNESIUM ION, Nuclear pore complex protein Nup50, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MVW
 
 | | Crystal structure of Chaetomium thermophilum Nup188 NTD (residues 1-1134) | | Descriptor: | GLYCEROL, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVT
 
 | | Crystal structure of the Chaetomium thermophilum Nup192-Nic96 complex (Nup192 residues 185-1756; Nic96 residues 187-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP192 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MVV
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup192-Nic96-Nup53-Nup145N complex (Nup192 residues 1-1756; Nic96 residues 240-301; Nup53 31-67; Nup145N 616-683) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP192, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MO5
 
 | | Crystal Structure of the ZnF4 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO1
 
 | | Crystal Structure of the ZnF1 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MVZ
 
 | | Single particle cryo-EM structure of the Chaetomium thermophilum Nup188-Nic96-Nup145N complex (Nup188 residues 1-1858; Nic96 residues 240-301; Nup145N residues 640-732) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP145N, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MW1
 
 | | Crystal structure of the Homo sapiens NUP93-NUP53 complex (NUP93 residues 174-819; NUP53 residues 84-150) | | Descriptor: | Nuclear pore complex protein Nup93, Nucleoporin Nup35 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7MNO
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) I656V mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.73 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNP
 
 | | Crystal Structure of the ZnF2 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
