3LE4
 
 | |
2LSL
 
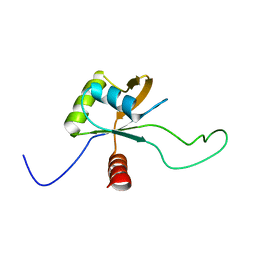 | | Solution structure of the C-terminal domain of Tetrahymena telomerase protein p65 | | Descriptor: | Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-05-01 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
2NTG
 
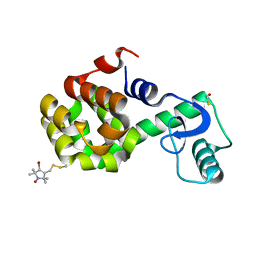 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R7 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(4-bromo-1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OU9
 
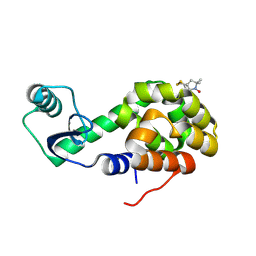 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1/R119A | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
3OEI
 
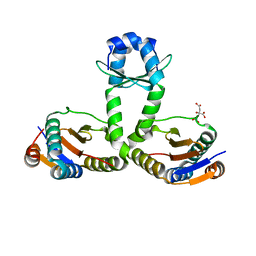 | | Crystal structure of Mycobacterium tuberculosis RelJK (Rv3357-Rv3358-RelBE3) | | Descriptor: | CITRATE ANION, RelJ (Antitoxin Rv3357), RelK (Toxin Rv3358) | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
2NTH
 
 | | Structure of Spin-labeled T4 Lysozyme Mutant L118R1 | | Descriptor: | Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2006-11-07 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2OU8
 
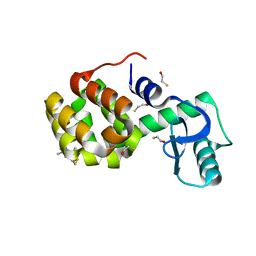 | | Structure of Spin-labeled T4 Lysozyme Mutant T115R1 at Room Temperature | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | EPR (1.8 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme.
Protein Sci., 16, 2007
|
|
2Q9D
 
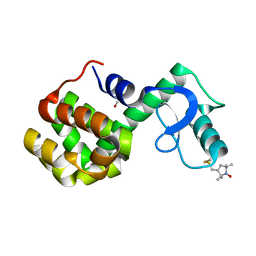 | | Structure of spin-labeled T4 lysozyme mutant A41R1 | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (1.4 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2Q9E
 
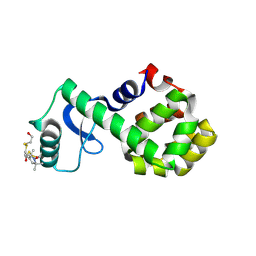 | | Structure of spin-labeled T4 lysozyme mutant S44R1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Lysozyme, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Guo, Z, Cascio, D, Hideg, K, Hubbell, W.L. | | Deposit date: | 2007-06-12 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | EPR (2.1 Å), X-RAY DIFFRACTION | | Cite: | Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme.
Protein Sci., 17, 2008
|
|
2MNW
 
 | |
1S3Q
 
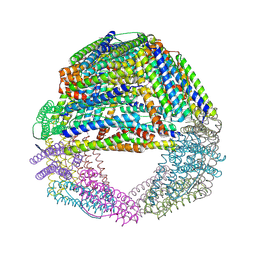 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus | | Descriptor: | ZINC ION, ferritin | | Authors: | Johnson, E, Cascio, D, Sawaya, M, Schroeder, I. | | Deposit date: | 2004-01-13 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
4I7A
 
 | | GrpN pentameric microcompartment shell protein from Rhodospirillum rubrum | | Descriptor: | CHLORIDE ION, Ethanolamine utilization protein EutN/carboxysome structural protein Ccml | | Authors: | Wheatley, N.M, Gidaniyan, S.D, Cascio, D, Yeates, T.O. | | Deposit date: | 2012-11-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins.
Protein Sci., 22, 2013
|
|
4IHW
 
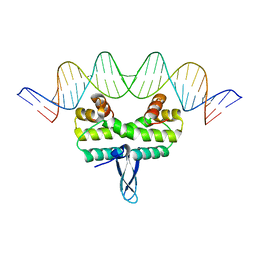 | |
4IHY
 
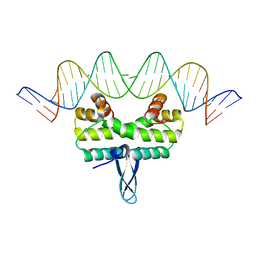 | |
4IHX
 
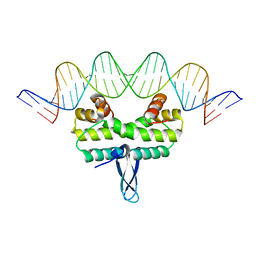 | |
4IHV
 
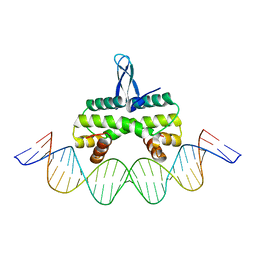 | |
1SQ3
 
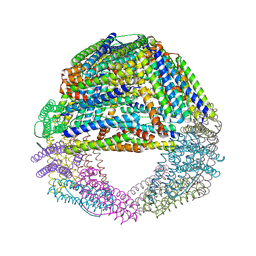 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus. | | Descriptor: | FE (III) ION, ferritin | | Authors: | Johnson, E, Cascio, D, Michael, S, Schroder, I. | | Deposit date: | 2004-03-17 | | Release date: | 2005-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
3EUD
 
 | | Structure of the CS domain of the essential H/ACA RNP assembly protein Shq1p | | Descriptor: | Protein SHQ1 | | Authors: | Singh, M, Cascio, D, Gonzales, F.A, Heckmann, N, Chanfreau, G, Feigon, J. | | Deposit date: | 2008-10-09 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Functional Studies of the CS Domain of the Essential H/ACA Ribonucleoparticle Assembly Protein SHQ1.
J.Biol.Chem., 284, 2009
|
|
3G3X
 
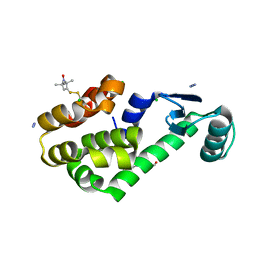 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 100 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3V
 
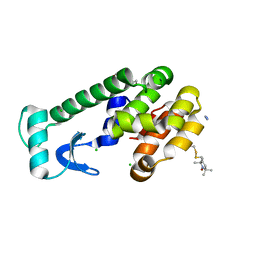 | | Crystal structure of spin labeled T4 Lysozyme (V131R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
3G3W
 
 | | Crystal structure of spin labeled T4 Lysozyme (T151R1) at 291 K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Hubbell, W.L. | | Deposit date: | 2009-02-02 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural origin of weakly ordered nitroxide motion in spin-labeled proteins.
Protein Sci., 18, 2009
|
|
5D4N
 
 | | Structure of CPII bound to ADP, AMP and acetate, from Thiomonas intermedia K12 | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
5E3L
 
 | |
5D4L
 
 | | Structure of the apo form of CPII from Thiomonas intermedia K12, a nitrogen regulatory PII-like protein | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-08-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
5E3M
 
 | | Crystal structure of Fis bound to 27bp DNA F35 (AAATTAGTTTGAATCTCGAGCTAATTT) | | Descriptor: | DNA (27-MER), DNA-binding protein Fis | | Authors: | Stella, S, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2015-10-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | DNA Sequence Determinants Controlling Affinity, Stability and Shape of DNA Complexes Bound by the Nucleoid Protein Fis.
Plos One, 11, 2016
|
|
