7PZ0
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 9.81x10^6 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYQ
 
 | | Structure of an LPMO (expressed in E.coli) at 6.35x10^6 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYY
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 5.05x10^5 Gy | | Descriptor: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYN
 
 | | Structure of an LPMO (expressed in E.coli) at 2.31x10^5 Gy | | Descriptor: | Auxiliary activity 9, COPPER (II) ION, SULFATE ION | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXS
 
 | | Room temperature X-ray structure of LPMO at 1.91x10^3 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYP
 
 | | Structure of an LPMO (expressed in E.coli) at 2.13x10^6 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXU
 
 | | LsAA9_A chemically reduced with ascorbic acid (low X-ray dose) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYL
 
 | | Structure of an LPMO (expressed in E.coli) at 1.49x10^4 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYX
 
 | | Structure of LPMO (expressed in E.coli) with cellotriose at 2.74x10^5 Gy | | Descriptor: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYM
 
 | | Structure of an LPMO (expressed in E.coli) at 5.61x10^4 Gy | | Descriptor: | Auxiliary activity 9, COPPER (II) ION, SULFATE ION | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYO
 
 | | Structure of an LPMO (expressed in E.coli) at 2.31x10^5 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYU
 
 | | Structure of an LPMO (expressed in E.coli) at 1.49x10^4 Gy | | Descriptor: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6K0J
 
 | | The co-crystal structure of DYRK2 with a small molecule inhibitor | | Descriptor: | 3-(2,7-dimethoxyacridin-9-yl)sulfanylpropan-1-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Tiantian, W, Xiao, J. | | Deposit date: | 2019-05-06 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Inhibition of dual-specificity tyrosine phosphorylation-regulated kinase 2 perturbs 26S proteasome-addicted neoplastic progression.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7LZ7
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5k | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
7LZ8
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5t | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[2-(ethylamino)pyrido[3,2-d]pyrimidin-4-yl]-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2021-03-09 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
4NYQ
 
 | | In-vivo crystallisation (midguts of a viviparous cockroach) and structure at 1.2 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Coussens, N.P, Gallat, F.-X, Ramaswamy, S, Yagi, K, Tobe, S.S, Stay, B, Chavas, L.M.G. | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
4NYR
 
 | | In-vivo crystallisation (midguts of a viviparous cockroach) and structure at 2.5 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Coussens, N.P, Gallat, F.-X, Ramaswamy, S, Yagi, K, Tobe, S.S, Stay, B, Chavas, L.M.G. | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
6X1C
 
 | | Tubulin-RB3_SLD-TTL in complex with compound 5j | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1F
 
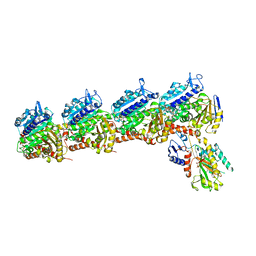 | | Tubulin-RB3_SLD-TTL in complex with compound 5m | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-methoxy-4-(2-methyl-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6X1E
 
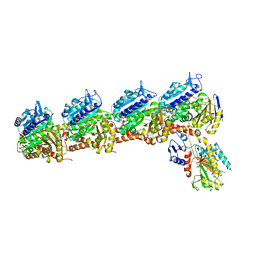 | | Tubulin-RB3_SLD-TTL in complex with compound 5l | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloro-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-18 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray Crystallography-Guided Design, Antitumor Efficacy, and QSAR Analysis of Metabolically Stable Cyclopenta-Pyrimidinyl Dihydroquinoxalinone as a Potent Tubulin Polymerization Inhibitor.
J.Med.Chem., 64, 2021
|
|
6BRF
 
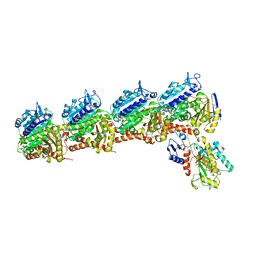 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[3,2-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-30 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BRY
 
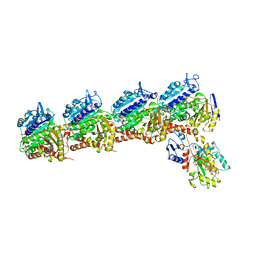 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 6a | | Descriptor: | 1-(2-chlorofuro[3,2-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BR1
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloro-4-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)pyrido[2,3-d]pyrimidine, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6BS2
 
 | | Tubulin-RB3_SLD-TTL in complex with heterocyclic pyrimidine compound 8b | | Descriptor: | 1-(3,6-dimethyl[1,2]oxazolo[5,4-d]pyrimidin-4-yl)-6-methoxy-1,2,3,4-tetrahydroquinoline, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2017-12-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Heterocyclic-Fused Pyrimidines as Novel Tubulin Polymerization Inhibitors Targeting the Colchicine Binding Site: Structural Basis and Antitumor Efficacy.
J. Med. Chem., 61, 2018
|
|
6DHK
 
 | | Bovine glutamate dehydrogenase complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation.
Biochemistry, 42, 2003
|
|
