2KBJ
 
 | | solution structure of BmKalphaTx11 (minor conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2KBH
 
 | | solution structure of BmKalphaTx11 (major conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2I0N
 
 | | Structure of Dictyostelium discoideum Myosin VII SH3 domain with adjacent proline rich region | | Descriptor: | Class VII unconventional myosin | | Authors: | Wang, Q, Deloia, M.A, Kang, Y, Litchke, C, Titus, M.A, Walters, K.J. | | Deposit date: | 2006-08-10 | | Release date: | 2007-01-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The SH3 domain of a M7 interacts with its C-terminal proline-rich region.
Protein Sci., 16, 2007
|
|
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR3
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4TTR
 
 | |
4TTQ
 
 | |
4TTP
 
 | |
6A0Q
 
 | | The crystal structure of Lpg2622_E64 complex | | Descriptor: | Lpg2622, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
6A0N
 
 | | The crystal structure of apo-Lpg2622 | | Descriptor: | Lpg2622 | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
2AST
 
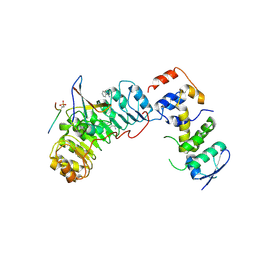 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
6UPL
 
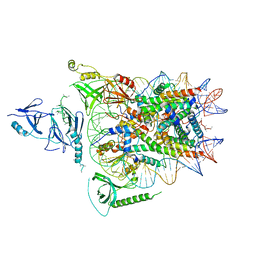 | | Structure of FACT_subnucleosome complex 2 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
6UPK
 
 | | Structure of FACT_subnucleosome complex 1 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
7CQQ
 
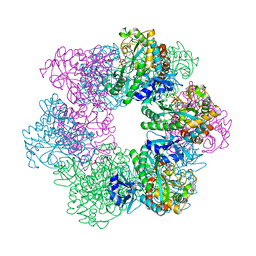 | | GmaS in complex with AMPPNP and MetSox | | Descriptor: | (2S)-2-AMINO-4-(METHYLSULFONIMIDOYL)BUTANOIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQL
 
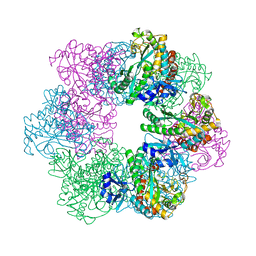 | | Apo GmaS without ligand | | Descriptor: | Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQN
 
 | | GmaS in complex with AMPPCP | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQU
 
 | | GmaS/ADP/MetSox-P complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQX
 
 | | GmaS/ADP complex-Conformation 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
7CQW
 
 | | GmaS/ADP complex-Conformation 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Type III glutamate--ammonia ligase | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-08-11 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structures of gamma-glutamylmethylamide synthetase provide insight into bacterial metabolism of oceanic monomethylamine.
J.Biol.Chem., 296, 2020
|
|
8Y74
 
 | |
4HXN
 
 | | Brd4 Bromodomain 1 complex with 4-(2-FLUOROPHENYL)-1,3-THIAZOL-2(3H)-ONE inhibitor | | Descriptor: | 4-(2-fluorophenyl)-1,3-thiazol-2(3H)-one, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXK
 
 | | Brd4 Bromodomain 1 complex with 6,7-DIHYDROTHIENO[3,2-C]PYRIDIN-5(4H)-YL(1H-IMIDAZOL-1-YL)METHANONE inhibitor | | Descriptor: | 6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl(1H-imidazol-1-yl)methanone, Bromodomain-containing protein 4 | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXR
 
 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]THIOPHENE-2-SULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]thiophene-2-sulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
4HXS
 
 | | Brd4 Bromodomain 1 complex with N-[3-(2-OXO-2,3-DIHYDRO-1,3-THIAZOL-4-YL)PHENYL]-1-PHENYLMETHANESULFONAMIDE inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]-1-phenylmethanesulfonamide | | Authors: | Chen, T.T, Cao, D.Y, Chen, W.Y, Xiong, B, Shen, J.K, Xu, Y.C. | | Deposit date: | 2012-11-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Drug Discovery of 2-Thiazolidinones as Inhibitors of the Histone Reader BRD4 Bromodomain.
J.Med.Chem., 56, 2013
|
|
