4P3O
 
 | | Structural Basis for Full-Spectrum Inhibition of Threonyl-tRNA Synthetase by Borrelidin 2 | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, GLYCEROL, Threonine--tRNA ligase, ... | | Authors: | Fang, P, Yu, X, Chen, K, Chen, X, Guo, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase.
Nat Commun, 6, 2015
|
|
6O6B
 
 | | Rotavirus A-VP3 (RVA-VP3) | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Protein VP3 | | Authors: | Kumar, D, Yu, X, Prasad, V, Wang, Z. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A sub-atomic resolution cryo-EM of full-length Rotavirus A-VP3 (RVA-VP3)
To Be Published
|
|
4P3N
 
 | | Structural Basis for Full-Spectrum Inhibition of Threonyl-tRNA Synthetase by Borrelidin 1 | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonine--tRNA ligase, cytoplasmic, ... | | Authors: | Fang, P, Yu, X, Chen, K, Chen, X, Guo, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase.
Nat Commun, 6, 2015
|
|
6OVX
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4QAP
 
 | | The second sphere residue T263 is important for function and activity of PTP1B through modulating WPD loop | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xiao, P, Wang, X, Wang, H.M, Fu, X.L, Cui, F.A, Yu, X, Bi, W.X, Sun, J.P. | | Deposit date: | 2014-05-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The second-sphere residue T263 is important for the function and catalytic activity of PTP1B via interaction with the WPD-loop
Int.J.Biochem.Cell Biol., 57, 2014
|
|
6OVQ
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | Descriptor: | GLYCEROL, Putative Side chain reductase | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OW0
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+ and PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MtmW, ... | | Authors: | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | Deposit date: | 2019-05-08 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
9DH3
 
 | | Cryo-EM structure of NLRP3 complex with Compound C | | Descriptor: | 2-[(4S)-5-ethyl-8-oxothieno[2',3':4,5]pyrrolo[1,2-d][1,2,4]triazin-7(8H)-yl]-N-(pyrimidin-4-yl)acetamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Matico, R, Grauwen, K, Van Gool, M, Muratore, E.M, Yu, X, Abdiaj, I, Adhikary, S, Adriaensen, I, Aranzazu, G.M, Alcazar, J, Bassi, M, Brisse, E, Canellas, S, Chaudhuri, S, Chauhan, D, Delgado, F, Dieguez-Vazquez, A, Du Jardin, M, Eastham, V, Finley, M, Jacobs, T, Keustermans, K, Kuhn, R, Llaveria, J, Leenaerts, J, Linares, M.L, Martin, M.L, Martinez, C, Miller, R, Munoz, F.M, Nooyens, A, Perez, L.B, Perrier, M, Pietrak, B, Serre, J, Sharma, S, Somers, M, Suarez, J, Tresadern, G, Trabanco, A.A, Van den Bulck, D, Van Hauwermeiren, F, Varghese, T, Vega, J.A, Youssef, S.A, Edwards, M.J, Oehlrich, D, Van Opdenbosch, N. | | Deposit date: | 2024-09-03 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Navigating from cellular phenotypic screen to clinical candidate: selective targeting of the NLRP3 inflammasome.
Embo Mol Med, 17, 2025
|
|
4X4J
 
 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
3KZC
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase | | Descriptor: | N-acetylornithine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
J.Biol.Chem., 280, 2005
|
|
3KZM
 
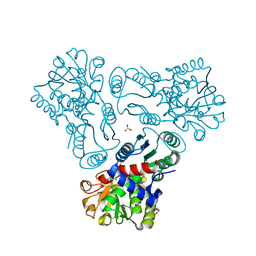 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KZK
 
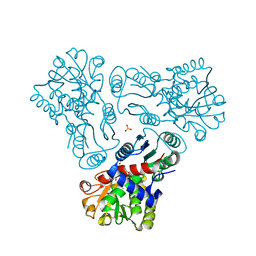 | | Crystal structure of acetylornithine transcarbamylase complexed with acetylcitrulline | | Descriptor: | (S)-2-ACETAMIDO-5-UREIDOPENTANOIC ACID, N-acetylornithine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
J.Biol.Chem., 280, 2005
|
|
3KZN
 
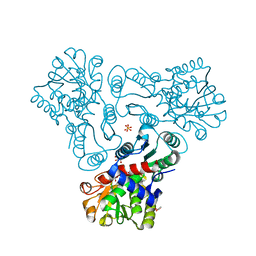 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with N-acetyl-L-ornirthine | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, N~2~-ACETYL-L-ORNITHINE, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
2FG6
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with sulfate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, SULFATE ION, putative ornithine carbamoyltransferase | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Mendel, T. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
3KZO
 
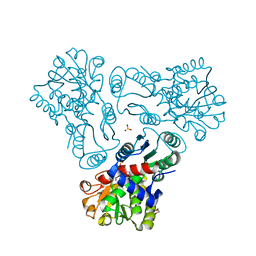 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate and N-acetyl-L-norvaline | | Descriptor: | GLYCEROL, N-ACETYL-L-NORVALINE, N-acetylornithine carbamoyltransferase, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3M4N
 
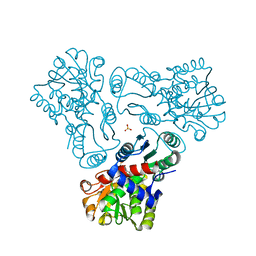 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302A mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5D
 
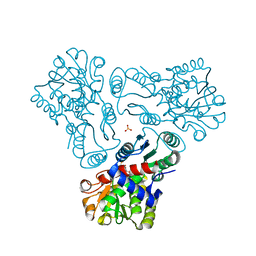 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302R mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5C
 
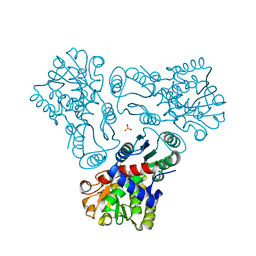 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3J89
 
 | | Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies | | Descriptor: | synthetic peptide | | Authors: | Egelman, E.H, Xu, C, DiMaio, F, Magnotti, E, Modlin, C, Yu, X, Wright, E, Baker, D, Conticello, V.P. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural plasticity of helical nanotubes based on coiled-coil assemblies.
Structure, 23, 2015
|
|
2FG7
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with carbamoyl phosphate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, SULFATE ION, ... | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Tuchman, M. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
3M4J
 
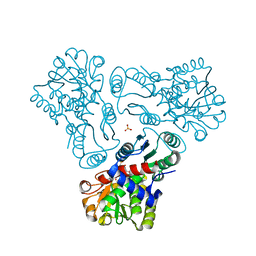 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
3J9R
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2HI5
 
 | | Model for bacteriophage fd from cryo-EM | | Descriptor: | Coat protein B | | Authors: | Wang, Y.A, Yu, X, Overman, S, Tsuboi, M, Thomas, G.J, Egelman, E.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The Structure of a Filamentous Bacteriophage
J.Mol.Biol., 361, 2006
|
|
3TXX
 
 | | Crystal structure of putrescine transcarbamylase from Enterococcus faecalis | | Descriptor: | Putrescine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Zhao, G, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-09-23 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of putrescine transcarbamylase from Enterococcus faecalis: Structural insights into the oligomeric assembly and the active site
To be Published
|
|
