8H00
 
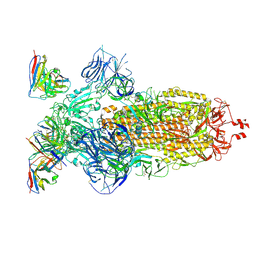 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
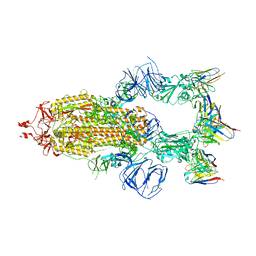 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
5XL1
 
 | |
5XLC
 
 | |
5XL3
 
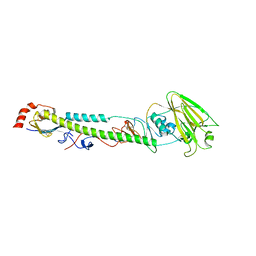 | | Complex structure of H4 hemagglutinin from avian influenza H4N6 virus with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Song, H, Qi, J, Gao, G.F. | | Deposit date: | 2017-05-10 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation by Influenza A Virus Hemagglutinin H4
Cell Rep, 20, 2017
|
|
5XL5
 
 | |
5WXD
 
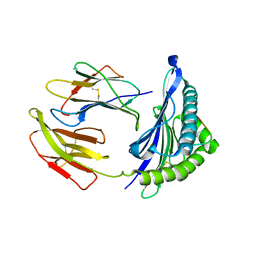 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 1) | | Descriptor: | Beta-2-microglobulin, H7-25, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5WXC
 
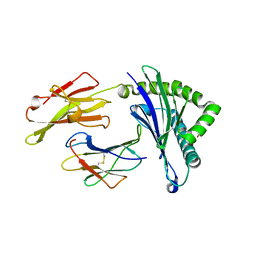 | | Crystal Structure of HLA-A*2402 in complex with avian influenza A(H7N9) virus-derived peptide H7-25 (data set 2) | | Descriptor: | Beta-2-microglobulin, H7-25-F, HLA class I histocompatibility antigen, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-07 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.295 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
5WWU
 
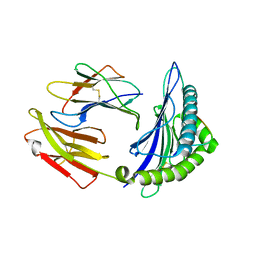 | | Crystal Structure of HLA-A*2402 in complex with 2009 pandemic influenza A(H1N1) virus and avian influenza A(H5N1) virus-derived peptide H1-25 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Zhao, M, Liu, K, Chai, Y, Qi, J, Liu, J, Gao, G.F. | | Deposit date: | 2017-01-05 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Heterosubtypic Protections against Human-Infecting Avian Influenza Viruses Correlate to Biased Cross-T-Cell Responses.
Mbio, 9, 2018
|
|
9JOD
 
 | | Capsid structure of Escherichia phage Mu | | Descriptor: | Major capsid protein | | Authors: | Zhou, J.Q, Liu, H.R. | | Deposit date: | 2024-09-24 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | In situ structures of the contractile nanomachine myophage Mu in both its extended and contracted states.
J.Virol., 99, 2025
|
|
5F94
 
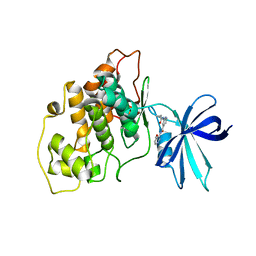 | | Crystal structure of GSK3b in complex with Compound 15: 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-methoxypyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
9KI1
 
 | | Baseplate structure of Escherichia phage Mu | | Descriptor: | Baseplate hub protein gp44, Baseplate protein gp46, Baseplate protein gp47, ... | | Authors: | Zhou, J.Q, Liu, H.R. | | Deposit date: | 2024-11-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structures of the contractile nanomachine myophage Mu in both its extended and contracted states.
J.Virol., 99, 2025
|
|
9KNU
 
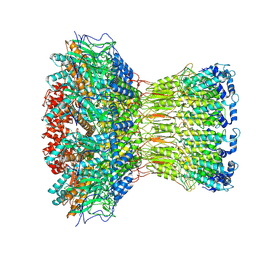 | |
9KHX
 
 | | Neck structure of Escherichia phage Mu | | Descriptor: | Gene product J, Portal protein | | Authors: | Zhou, J.Q, Liu, H.R. | | Deposit date: | 2024-11-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of the contractile nanomachine myophage Mu in both its extended and contracted states.
J.Virol., 99, 2025
|
|
9KHY
 
 | | Terminator and trunk structure of Escherichia phage Mu | | Descriptor: | Probable tail terminator protein, Tail sheath protein, Tail tube protein | | Authors: | Zhou, J.Q, Liu, H.R. | | Deposit date: | 2024-11-11 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of the contractile nanomachine myophage Mu in both its extended and contracted states.
J.Virol., 99, 2025
|
|
9LJ8
 
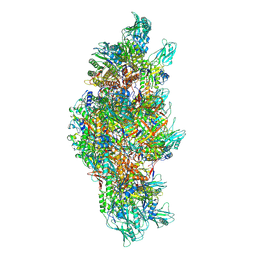 | | Tail structure of bacteriophage Mu in contracted state | | Descriptor: | Probable tail terminator protein, Tail sheath protein, Tail tube protein | | Authors: | Liu, H.R, Zhou, J.Q. | | Deposit date: | 2025-01-14 | | Release date: | 2025-02-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | In situ structures of the contractile nanomachine myophage Mu in both its extended and contracted states.
J.Virol., 99, 2025
|
|
5F95
 
 | | Crystal structure of GSK3b in complex with Compound 18: 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide | | Descriptor: | 2-[(cyclopropylcarbonyl)amino]-N-(4-phenylpyridin-3-yl)pyridine-4-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Kish, K, Luo, G, Dubowchick, G.M. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.525 Å) | | Cite: | Discovery of Isonicotinamides as Highly Selective, Brain Penetrable, and Orally Active Glycogen Synthase Kinase-3 Inhibitors.
J.Med.Chem., 59, 2016
|
|
5HGU
 
 | |
4QNX
 
 | | Crystal structure of apo-CmoB | | Descriptor: | SULFATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
4QNV
 
 | | Crystal structure of Cx-SAM bound CmoB from E. coli in P6122 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
4QNU
 
 | | Crystal structure of CmoB bound with Cx-SAM in P21212 | | Descriptor: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, PHOSPHATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Kim, J, Toro, R, Bhosle, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-06-18 | | Release date: | 2014-09-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of the CmoB carboxymethyl transferase utilized for selective tRNA wobble modification.
Nucleic Acids Res., 43, 2015
|
|
5ZZ3
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 | | Descriptor: | Butyrophilin, subfamily 3, member A3 isoform b variant | | Authors: | Yang, Y.Y, Li, X, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
5ZXK
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A1 | | Authors: | Yang, Y.Y, Liu, W.D, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6IW4
 
 | |
