8WS3
 
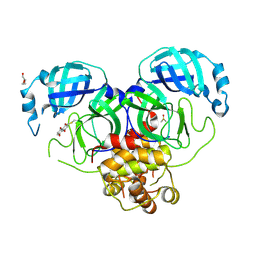 | | Crystal structure of SARS-CoV-2 Main Protease (Mpro) with covalent inhibitor 5,8-Dihydroxy-1,4-naphthoquinone | | Descriptor: | 3C-like proteinase nsp5, 5,8-bis(oxidanyl)naphthalene-1,4-dione, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Wu, D. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Covalent inhibitors of SARS-CoV-2 main protease
To Be Published
|
|
4PW8
 
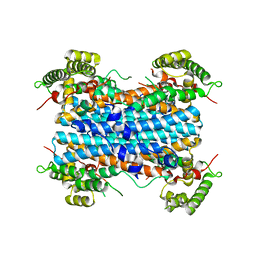 | | Human tryptophan 2,3-dioxygenase | | Descriptor: | COBALT (II) ION, Tryptophan 2,3-dioxygenase | | Authors: | Meng, B, Wu, D, Gu, J.H, Ouyang, S.Y, Ding, W, Liu, Z.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and functional analyses of human tryptophan 2,3-dioxygenase
Proteins, 82, 2014
|
|
8XKF
 
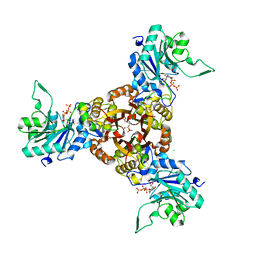 | | Crystal structure of Helicobacter pylori IspDF with substrate CTP | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XKG
 
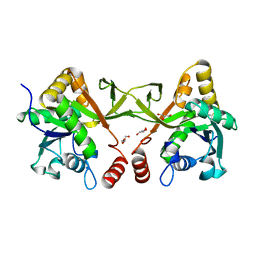 | | Crystal structure of Acinetobacter baumannii IspD | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, GLYCEROL | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-23 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XHU
 
 | | Crystal structure of Helicobacter pylori IspDF | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional enzyme IspD/IspF, CHLORIDE ION, ... | | Authors: | Chen, X, Wu, D. | | Deposit date: | 2023-12-18 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two natural compounds as potential inhibitors against the Helicobacter pylori and Acinetobacter baumannii IspD enzymes.
Int J Antimicrob Agents, 63, 2024
|
|
8XSB
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Indirubin | | Descriptor: | (3~{Z})-3-(3-oxidanylidene-1~{H}-indol-2-ylidene)-1~{H}-indol-2-one, Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS6
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Tapinarof | | Descriptor: | 5-[(E)-2-phenylethenyl]-2-propan-2-yl-benzene-1,3-diol, Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS7
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with FICZ | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5,11-dihydroindolo[3,2-b]carbazole-12-carbaldehyde, Aryl hydrocarbon receptor, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS9
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with beta-Naphthoflavone | | Descriptor: | (3R)-3-phenyl-2,3-dihydrobenzo[f]chromen-1-one, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Aryl hydrocarbon receptor, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XS8
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Benzo[a]pyrene | | Descriptor: | Aryl hydrocarbon receptor, Aryl hydrocarbon receptor nuclear translocator, DNAF, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
8XSA
 
 | | Crystal structure of the DNA-bound AHR-ARNT heterodimer in complex with Indigo | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(3-oxidanyl-1H-indol-2-yl)indol-3-one, Aryl hydrocarbon receptor, ... | | Authors: | Diao, X, Shang, Q, Wu, D. | | Deposit date: | 2024-01-09 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Structural basis for the ligand-dependent activation of heterodimeric AHR-ARNT complex.
Nat Commun, 16, 2025
|
|
4RHW
 
 | | Crystal structure of Apaf-1 CARD and caspase-9 CARD complex | | Descriptor: | Apoptotic protease-activating factor 1, CHLORIDE ION, Caspase-9, ... | | Authors: | Hu, Q, Wu, D, Yan, C, Shi, Y. | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
4TZ7
 
 | | Crystal structure of type I phosphatidylinositol 4-phosphate 5-kinase alpha from Zebrafish | | Descriptor: | Phosphatidylinositol-4-phosphate 5-kinase, type I, alpha | | Authors: | Hu, J, Qin, Y, Wang, J, Li, L, Wu, D, Ha, Y. | | Deposit date: | 2014-07-09 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled.
Nat Commun, 6, 2015
|
|
7W80
 
 | | Crystal Structure of the Heterodimeric HIF-2 in Complex with Antagonist Belzutifan | | Descriptor: | 3-{[(1S,2S,3R)-2,3-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Ren, X, Diao, X, Zhuang, J, Wu, D. | | Deposit date: | 2021-12-07 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.754 Å) | | Cite: | Structural basis for the allosteric inhibition of hypoxia-inducible factor (HIF)-2 by belzutifan.
Mol.Pharmacol., 2022
|
|
4FC6
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | HEXANOYL-COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
4FC7
 
 | | Studies on DCR shed new light on peroxisomal beta-oxidation: Crystal structure of the ternary complex of pDCR | | Descriptor: | COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Peroxisomal 2,4-dienoyl-CoA reductase | | Authors: | Hua, T, Wu, D, Wang, J, Shaw, N, Liu, Z.-J. | | Deposit date: | 2012-05-24 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Studies of human 2,4-dienoyl CoA reductase shed new light on peroxisomal beta-oxidation of unsaturated fatty acids
J.Biol.Chem., 287, 2012
|
|
6KUY
 
 | | Crystal structure of the alpha2A adrenergic receptor in complex with a partial agonist | | Descriptor: | (2~{S})-4-fluoranyl-2-(1~{H}-imidazol-5-yl)-1-propan-2-yl-2,3-dihydroindole, Alpha2A adrenergic receptor, DI(HYDROXYETHYL)ETHER | | Authors: | Qu, L, Zhou, Q.T, Wu, D, Zhao, S.W. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the alpha2A adrenergic receptor in complex with a partial agonist
To Be Published
|
|
7V7L
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7V7W
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
7C1S
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA and Leu-SNAC | | Descriptor: | Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A, S-(2-acetamidoethyl) (2S)-2-azanyl-4-methyl-pentanethioate | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1L
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1U
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140V/R148A in a "product-released" conformation | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1H
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1R
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1P
 
 | | Crystal structure of the starter condensation domain of the rhizomide synthetase RzmA mutant H140V, R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
