8XHP
 
 | | The apo structure of SsBcmC | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Fe/2OG dependent dioxygenase, ... | | Authors: | Wu, L, Tang, G.L, Zhou, J.H. | | Deposit date: | 2023-12-18 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.86000824 Å) | | Cite: | Three distinct strategies lead to programmable aliphatic C-H oxidation in bicyclomycin biosynthesis.
Nat Commun, 16, 2025
|
|
8XHT
 
 | | The apo structure of PaBcmG | | Descriptor: | 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | Authors: | Wu, L, Tang, G.L, Zhou, J.H. | | Deposit date: | 2023-12-18 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.81953633 Å) | | Cite: | Three distinct strategies lead to programmable aliphatic C-H oxidation in bicyclomycin biosynthesis.
Nat Commun, 16, 2025
|
|
8XHY
 
 | | The complex structure of mutant T307A of SsBcmE and its natural substrate cIL | | Descriptor: | (3~{S},6~{S})-3-[(2~{S})-butan-2-yl]-6-(2-methylpropyl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Tang, G.L, Zhou, J.H. | | Deposit date: | 2023-12-18 | | Release date: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.71001971 Å) | | Cite: | Three distinct strategies lead to programmable aliphatic C-H oxidation in bicyclomycin biosynthesis.
Nat Commun, 16, 2025
|
|
7FE5
 
 | |
7FE6
 
 | |
7FE0
 
 | |
7V3E
 
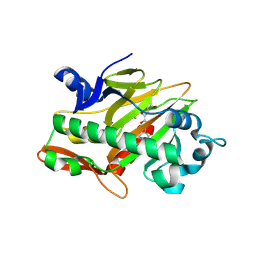 | | The complex structure of soBcmB and its intermediate product 1a | | Descriptor: | (3S,6Z)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2X
 
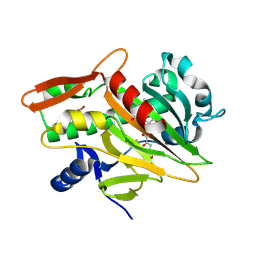 | | The complex structure of soBcmB and its substrate 1 | | Descriptor: | (3S,6S)-3-[(2S)-butan-2-yl]-6-[(2R)-2-methyl-2,3-bis(oxidanyl)propyl]piperazine-2,5-dion, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.08387423 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V3O
 
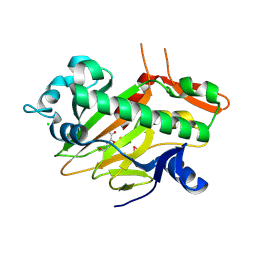 | | The structure of Se-SoBcmB with Fe(II)and AKG | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.83004665 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V2U
 
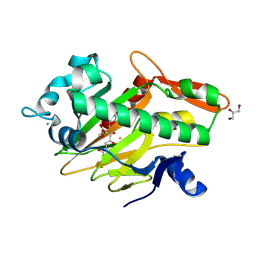 | | The complex structure of SoBcmB and its product 2f | | Descriptor: | (1S,5S,6S)-5-methyl-1-[(1S,2S)-2-methyl-1,2,3-tris(oxidanyl)propyl]-2-oxa-7,9-diazabicyclo[4.2.2]decane-8,10-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.00009918 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V34
 
 | | The complex structure of soBcmB and its product 1d | | Descriptor: | (3S,4S,5S,8S)-8-[(2S)-butan-2-yl]-3-methyl-3,4-bis(oxidanyl)-1-oxa-7,10-diazaspiro[4.5]decane-6,9-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.000169 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7V36
 
 | | The complex structure of soBcmB and its intermediate product 2a | | Descriptor: | (3Z,6S)-3-[(2R)-2-methyl-2,3-bis(oxidanyl)propylidene]-6-[(2S)-4-oxidanylbutan-2-yl]piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.96009171 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
7WJ6
 
 | |
7WJ7
 
 | |
7V3N
 
 | | The complex structure of soBcmB-D307A and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, FE (II) ION, ... | | Authors: | Wu, L, Zhou, J.H. | | Deposit date: | 2021-08-10 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.850011 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
5HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
4HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, CHAIN HA1, ... | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
3HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
1HTM
 
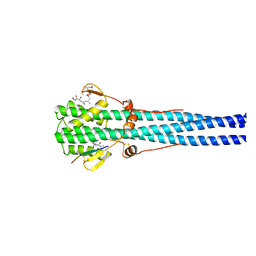 | | STRUCTURE OF INFLUENZA HAEMAGGLUTININ AT THE PH OF MEMBRANE FUSION | | Descriptor: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN | | Authors: | Bullough, P.A, Hughson, F.M, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza haemagglutinin at the pH of membrane fusion.
Nature, 371, 1994
|
|
2HMG
 
 | | REFINEMENT OF THE INFLUENZA VIRUS HEMAGGLUTININ BY SIMULATED ANNEALING | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ (HA1 CHAIN), HEMAGGLUTININ (HA2 CHAIN), ... | | Authors: | Weis, W.I, Bruenger, A.T, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1989-09-11 | | Release date: | 1991-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refinement of the influenza virus hemagglutinin by simulated annealing.
J.Mol.Biol., 212, 1990
|
|
5JLG
 
 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and compound I, a piano-stool organometallic Ru(II) arene compound containing an O,S-chelating ligand | | Descriptor: | DIMETHYL SULFOXIDE, RUTHENIUM ION, Ribonuclease pancreatic, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(ii) piano-stool compound containing an O,S-chelating ligand.
Dalton Trans, 45, 2016
|
|
4V9I
 
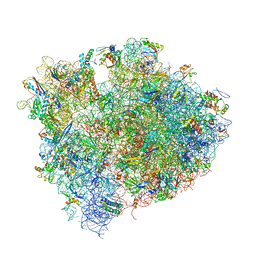 | | Crystal structure of thermus thermophilus 70S in complex with tRNAs and mRNA containing a pseudouridine in a stop codon | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S Ribosomal protein S10, ... | | Authors: | Fernandez, I.S, Ng, C.L, Kelley, A.C, Guowei, W, Yu, Y.T, Ramakrishnan, V. | | Deposit date: | 2013-04-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unusual base pairing during the decoding of a stop codon by the ribosome.
Nature, 500, 2013
|
|
6XDB
 
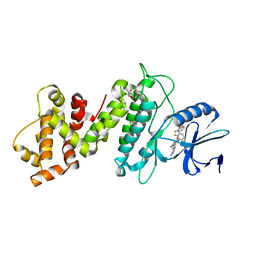 | | Crystal structure of IRE1a in complex with G-6904 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-N-(6-chloro-2-fluoro-3-{[(2-fluorophenyl)sulfonyl]amino}phenyl)-6-(1,3-dimethyl-1H-pyrazol-4-yl)quinazoline-8-carboxamide, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Wallweber, H.A, Weiru, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of BRaf-Sparing Amino-Thienopyrimidines with Potent IRE1 alpha Inhibitory Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
1ONI
 
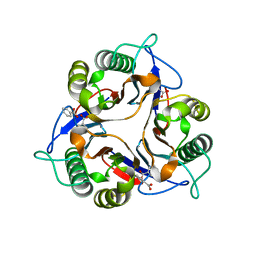 | | Crystal structure of a human p14.5, a translational inhibitor reveals different mode of ligand binding near the invariant residues of the Yjgf/UK114 protein family | | Descriptor: | 14.5 kDa translational inhibitor protein, BENZOIC ACID | | Authors: | Manjasetty, B.A, Delbrueck, H, Mueller, U, Erdmann, M.F, Heinemann, U. | | Deposit date: | 2003-02-28 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Homo sapiens protein hp14.5.
Proteins, 54, 2004
|
|
