4D4F
 
 | | Mutant P250A of bacterial chalcone isomerase from Eubacterium ramulus | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL | | Authors: | Thomsen, M, Kratzat, H, Hinrichs, W. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus.
Molecules, 27, 2022
|
|
3FPS
 
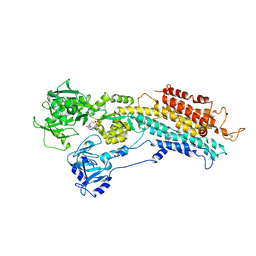 | | The Structure of Sarcoplasmic Reticulum Ca2+-ATPase Bound To Cyclopiazonic and ADP | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moncoq, K, Morth, J.P, Bublitz, M, Laursen, M, Nissen, P, Young, H.S. | | Deposit date: | 2009-01-06 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cyclopiazonic acid is complexed to a divalent metal ion when bound to the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 284, 2009
|
|
3FPB
 
 | | The Structure of Sarcoplasmic Reticulum Ca2+-ATPase Bound To Cyclopiazonic acid with ATP | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moncoq, K, Morth, J.P, Bublitz, M, Laursen, M, Nissen, P, Young, H.S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Cyclopiazonic acid is complexed to a divalent metal ion when bound to the sarcoplasmic reticulum Ca2+-ATPase.
J.Biol.Chem., 284, 2009
|
|
2YJH
 
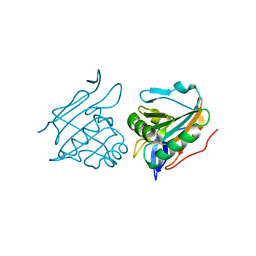 | | Thiol Peroxidase from Yersinia Psuedotuberculosis, inactive mutant C61S | | Descriptor: | THIOL PEROXIDASE | | Authors: | Beckham, K.S.H, Gabrielsen, M, Wang, D, Roe, A.J. | | Deposit date: | 2011-05-19 | | Release date: | 2012-03-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Characterisation of Tpx from Yersinia Pseudotuberculosis Reveals Insights Into the Binding of Salicylidene Acylhydrazide Compounds.
Plos One, 7, 2012
|
|
3ZPH
 
 | |
287D
 
 | |
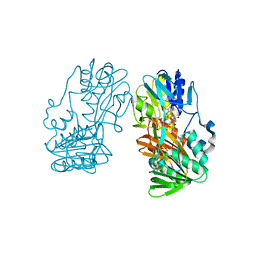
2GR0
 
 | |
2GQW
 
 | | Crystal structure of Ferredoxin reductase, BphA4 (oxidized form) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2006-04-22 | | Release date: | 2007-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2GR1
 
 | |
2GR3
 
 | |
2GR2
 
 | |
1HG9
 
 | | Solution structure of DNA:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*TP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-13 | | Release date: | 2002-01-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity.
Chemistry, 6, 2000
|
|
1O73
 
 | | Tryparedoxin from Trypanosoma brucei | | Descriptor: | TRYPAREDOXIN | | Authors: | Gabrielsen, M, Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-10-23 | | Release date: | 2003-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
3KM9
 
 | | Structure of complement C5 in complex with the C-terminal beta-grasp domain of SSL7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Laursen, N.S, Gordon, N, Hermans, S, Lorenz, N, Jackson, N, Wines, B, Spillner, E, Christensen, J.B, Jensen, M, Fredslund, F, Bjerre, M, Sottrup-Jensen, L, Fraser, J.D, Andersen, G.R. | | Deposit date: | 2009-11-10 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4F4O
 
 | | Structure of the Haptoglobin-Haemoglobin Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Andersen, C.B.F, Torvund-Jensen, M, Nielsen, M.J, Oliveira, C.L.P, Hersleth, H.P, Andersen, N.H, Pedersen, J.S, Andersen, G.R, Moestrup, S.K. | | Deposit date: | 2012-05-11 | | Release date: | 2012-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the haptoglobin-haemoglobin complex.
Nature, 489, 2012
|
|
7D6Q
 
 | | Crystal structure of the Stx2a | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Shiga toxin 2 B subunit, rRNA N-glycosylase | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
7D6R
 
 | | Crystal structure of the Stx2a complexed with MMA betaAla peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, MMA betaAla peptide, Shiga toxin 2 B subunit, ... | | Authors: | Takahashi, M, Tamada, M, Hibino, M, Senda, M, Okuda, A, Miyazawa, A, Senda, T, Nishikawa, K. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a peptide motif that potently inhibits two functionally distinct subunits of Shiga toxin.
Commun Biol, 4, 2021
|
|
3KQ4
 
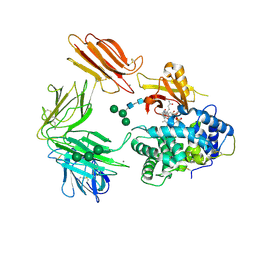 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
2G9Q
 
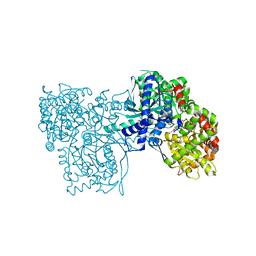 | | The crystal structure of the glycogen phosphorylase b- 1AB complex | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-D-ARABINITOL, Glycogen phosphorylase, muscle form, ... | | Authors: | Oikonomakos, N.G, Tiraidis, C, Leonidas, D.D, Zographos, S.E, Kristiansen, M, Agius, L. | | Deposit date: | 2006-03-07 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iminosugars as potential inhibitors of glycogenolysis: structural insights into the molecular basis of glycogen phosphorylase inhibition.
J.Med.Chem., 49, 2006
|
|
8S03
 
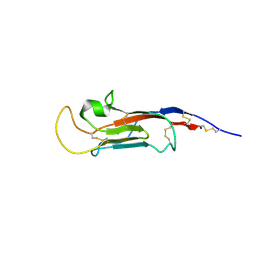 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep, 43, 2024
|
|
4WJG
 
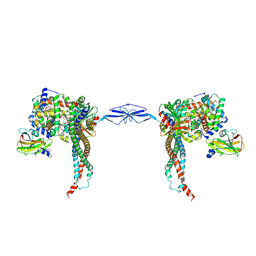 | | Structure of T. brucei haptoglobin-hemoglobin receptor binding to human haptoglobin-hemoglobin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin, ... | | Authors: | Stoedkilde, K, Torvund-Jensen, M, Moestrup, S.K, Andersen, C.B.F. | | Deposit date: | 2014-09-30 | | Release date: | 2014-11-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for trypanosomal haem acquisition and susceptibility to the host innate immune system.
Nat Commun, 5, 2014
|
|
3UTZ
 
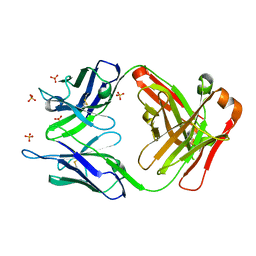 | | Endogenous-like inhibitory antibodies targeting activated metalloproteinase motifs show therapeutic potential | | Descriptor: | Metalloproteinase, heavy chain, light chain, ... | | Authors: | Sela-Passwell, N, Kikkeri, R, Dym, O, Rozenberg, H, Margalit, R, Arad-Yellin, R, Eisenstein, M, Brenner, O, Shoham, T, Danon, T, Shanzer, A, Sagi, I, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-11-27 | | Release date: | 2011-12-14 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential.
NAT.MED. (N.Y.), 18, 2012
|
|
4V3K
 
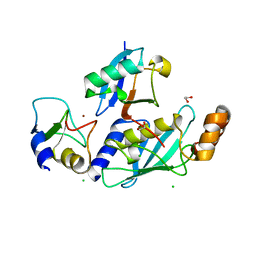 | | RNF38-UbcH5B-UB complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE RNF38, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
4V3L
 
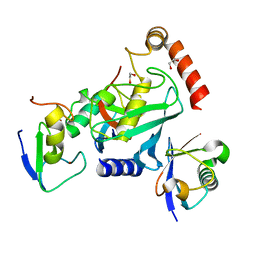 | | RNF38-UB-UbcH5B-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF38, POLYUBIQUITIN-C, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
