2VHH
 
 | | Crystal structure of a pyrimidine degrading enzyme from Drosophila melanogaster | | Descriptor: | CG3027-PA | | Authors: | Lundgren, S, Lohkamp, B, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-11-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of Beta-Alanine Synthase from Drosophila Melanogaster Reveals a Homooctameric Helical Turn-Like Assembly.
J.Mol.Biol., 377, 2008
|
|
1ELA
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-prolinamide, ACETIC ACID, CALCIUM ION, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
2VP6
 
 | |
1ELB
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-leucinamide, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1Z5R
 
 | | Crystal Structure of Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-03-18 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1ZG9
 
 | | Crystal Structure of 5-{[amino(imino)methyl]amino}-2-(sulfanylmethyl)pentanoic acid Bound to Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | 5-{[AMINO(IMINO)METHYL]AMINO}-2-(SULFANYLMETHYL)PENTANOIC ACID, ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-04-20 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1ZT1
 
 | | crystal structure of class I MHC H-2Kk in complex with an octapeptide | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, H-2 class I histocompatibility antigen, ... | | Authors: | Kellenberger, C, Roussel, A, Malissen, B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The H-2Kk MHC peptide-binding groove anchors the backbone of an octameric antigenic peptide in an unprecedented mode.
J Immunol., 175, 2005
|
|
1ZT7
 
 | | crystal structure of class I MHC H-2Kk in complex with a nonapeptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-K alpha chain, ... | | Authors: | Kellenberger, C, Roussel, A, Malissen, B. | | Deposit date: | 2005-05-26 | | Release date: | 2005-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The H-2Kk MHC peptide-binding groove anchors the backbone of an octameric antigenic peptide in an unprecedented mode.
J Immunol., 175, 2005
|
|
2V8D
 
 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-ALANINE SYNTHASE, ZINC ION | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8H
 
 | | Crystal structure of mutant E159A of beta-alanine synthase from Saccharomyces kluyveri in complex with its substrate N-carbamyl-beta- alanine | | Descriptor: | BETA-ALANINE SYNTHASE, BICINE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
3WGV
 
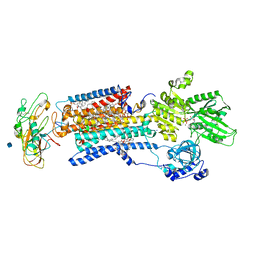 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state with oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
2V8G
 
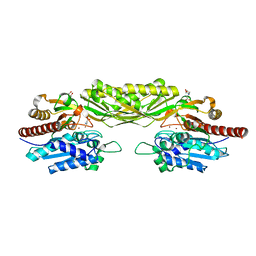 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri in complex with the product beta-alanine | | Descriptor: | BETA-ALANINE, BETA-ALANINE SYNTHASE, BICINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
2V8V
 
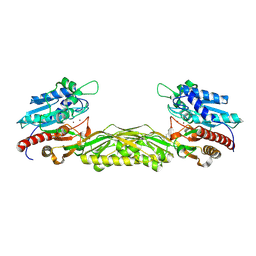 | | Crystal structure of mutant R322A of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-ALANINE SYNTHASE, N-(AMINOCARBONYL)-BETA-ALANINE, ... | | Authors: | Lundgren, S, Andersen, B, Piskur, J, Dobritzsch, D. | | Deposit date: | 2007-08-15 | | Release date: | 2007-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Yeast -Alanine Synthase Complexes Reveal the Mode of Substrate Binding and Large Scale Domain Closure Movements.
J.Biol.Chem., 282, 2007
|
|
3WGU
 
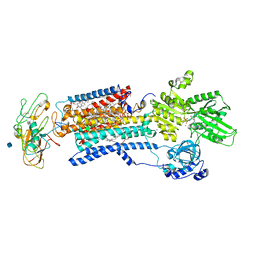 | | Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state without oligomycin | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kanai, R, Ogawa, H, Vilsen, B, Cornelius, F, Toyoshima, C. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a Na1-bound Na1,K1-ATPase preceding the E1P state
Nature, 502, 2013
|
|
1NAM
 
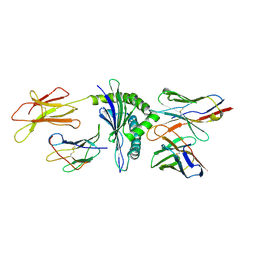 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BM3.3 T Cell Receptor alpha-Chain, BM3.3 T Cell Receptor beta-Chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1WYX
 
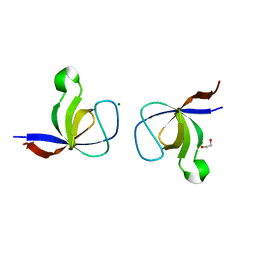 | | The Crystal Structure of the p130Cas SH3 Domain at 1.1 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CRK-associated substrate, MAGNESIUM ION | | Authors: | Wisniewska, M, Bossenmaier, B, Georges, G, Hesse, F, Dangl, M, Kuenkele, K.P, Ioannidis, I, Huber, R, Engh, R.A. | | Deposit date: | 2005-02-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | The 1.1 A resolution crystal structure of the p130cas SH3 domain and ramifications for ligand selectivity
J.Mol.Biol., 347, 2005
|
|
1OE0
 
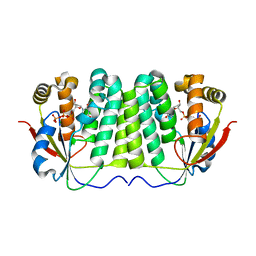 | | CRYSTAL STRUCTURE OF DROSOPHILA DEOXYRIBONUCLEOSIDE KINASE IN COMPLEX WITH DTTP | | Descriptor: | DEOXYRIBONUCLEOSIDE KINASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-17 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway:Studies of the Drosophila Deoxyribonucleoside Kinase
Biochemistry, 42, 2003
|
|
1OT3
 
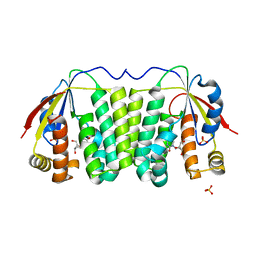 | | Crystal structure of Drosophila deoxyribonucleotide kinase complexed with the substrate deoxythymidine | | Descriptor: | Deoxyribonucleoside Kinase, SULFATE ION, THYMIDINE | | Authors: | Mikkelsen, N.E, Johansson, K, Karlsson, A, Knecht, W, Andersen, G, Piskur, J, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2003-03-21 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Feedback Inhibition of the Deoxyribonucleoside Salvage Pathway: Studies of the Drosophila Deoxyribonucleoside Kinase.
Biochemistry, 42, 2003
|
|
1OO3
 
 | |
1NAN
 
 | | MCH CLASS I H-2KB MOLECULE COMPLEXED WITH PBM1 PEPTIDE | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Reiser, J.-B, Darnault, C, Gregoire, C, Mosser, T, Mazza, G, Kearnay, A, van der Merwe, P.A, Fontecilla-Camps, J.C, Housset, D, Malissen, B. | | Deposit date: | 2002-11-28 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDR3 loop flexibility contributes to the degeneracy of TCR recognition
Nat.Immunol., 4, 2003
|
|
1OO4
 
 | |
1HRV
 
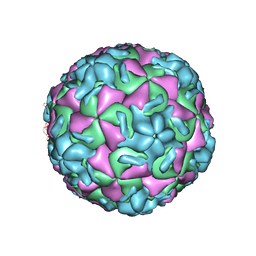 | | HRV14/SDZ 35-682 COMPLEX | | Descriptor: | 1-[2-HYDROXY-3-(4-CYCLOHEXYL-PHENOXY)-PROPYL]-4-(2-PYRIDYL)-PIPERAZINE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1995-03-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | SDZ 35-682, a new picornavirus capsid-binding agent with potent antiviral activity.
Antiviral Res., 26, 1995
|
|
8AW2
 
 | | Mouse serotonin 5-HT3A receptor in complex with vortioxetine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Lopez-Sanchez, U, Nury, H. | | Deposit date: | 2022-08-29 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural determinants for activity of the antidepressant vortioxetine at human and rodent 5-HT 3 receptors.
Nat.Struct.Mol.Biol., 2024
|
|
8AXD
 
 | | Human serotonin 5-HT3A receptor (apo, resting conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A | | Authors: | Lopez-Sanchez, U, Nury, H. | | Deposit date: | 2022-08-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural determinants for activity of the antidepressant vortioxetine at human and rodent 5-HT 3 receptors.
Nat.Struct.Mol.Biol., 2024
|
|
6U57
 
 | |
