5K7T
 
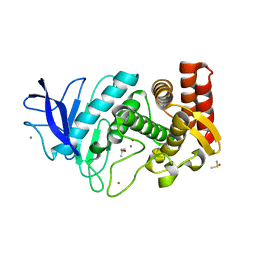 | | MicroED structure of thermolysin at 2.5 A resolution | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7O
 
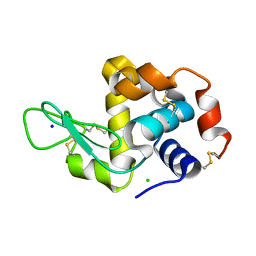 | | MicroED structure of lysozyme at 1.8 A resolution | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7N
 
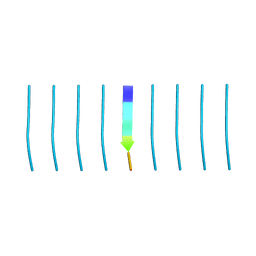 | | MicroED structure of tau VQIVYK peptide at 1.1 A resolution | | Descriptor: | VQIVYK | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2024-02-28 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.1 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5K7S
 
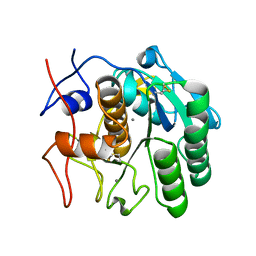 | | MicroED structure of proteinase K at 1.6 A resolution | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | de la Cruz, M.J, Hattne, J, Shi, D, Seidler, P, Rodriguez, J, Reyes, F.E, Sawaya, M.R, Cascio, D, Eisenberg, D, Gonen, T. | | Deposit date: | 2016-05-26 | | Release date: | 2017-04-05 | | Last modified: | 2018-08-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.6 Å) | | Cite: | Atomic-resolution structures from fragmented protein crystals with the cryoEM method MicroED.
Nat. Methods, 14, 2017
|
|
5KO0
 
 | | Human Islet Amyloid Polypeptide Segment 15-FLVHSSNNFGA-25 Determined by MicroED | | Descriptor: | THIOCYANATE ION, hIAPP(15-25)WT | | Authors: | Krotee, P.A.L, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Shi, D, Nannenga, B.L, Hattne, J, Reyes, F.E, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2016-06-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity.
Elife, 6, 2017
|
|
8FHJ
 
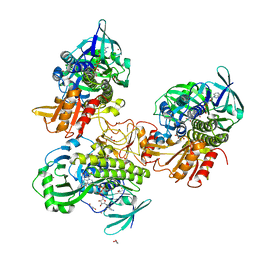 | | Crystal structure of a FAD monooxygenease from Methylocystis sp. Strain SB2 | | Descriptor: | BROMIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Stewart, A.M, Sawaya, M.R, Stewart, C.E. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of MbnF: an NADPH-dependent flavin monooxygenase from Methylocystis strain SB2.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8G3K
 
 | |
8G42
 
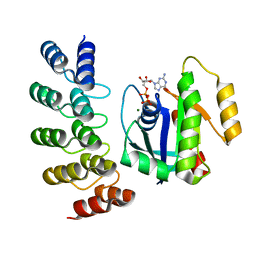 | | KRAS G12C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G2V
 
 | | Cryo-EM structure of recombinant human LECT2 amyloid fibril core | | Descriptor: | Leukocyte cell-derived chemotaxin-2 | | Authors: | Richards, L.S, Flores, M.D, Zink, S, Schibrowsky, N.A, Sawaya, M.R, Rodriguez, J.A. | | Deposit date: | 2023-02-06 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.715 Å) | | Cite: | Cryo-EM structure of a human LECT2 amyloid fibril reveals a network of polar ladders at its core.
Structure, 31, 2023
|
|
8FQ7
 
 | | Nanobody with WIW inserted in CDR3 loop to Inhibit Growth of Alzheimer's Tau fibrils | | Descriptor: | GLYCEROL, Nanobody with WIW insert in CDR3 loop to target tau fibrils | | Authors: | Abskharon, R, Sawaya, M.R, Cascio, D.C, Eisenberg, D.S. | | Deposit date: | 2023-01-05 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of nanobodies that inhibit seeding of Alzheimer's patient-extracted tau fibrils.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G47
 
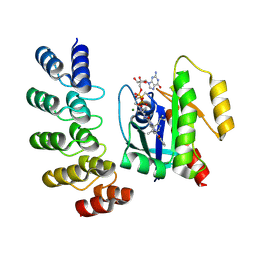 | | KRAS G12C complex with GDP and AMG 510 imaged on a cryo-EM imaging scaffold | | Descriptor: | AMG 510 (bound form), GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-08 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8G4H
 
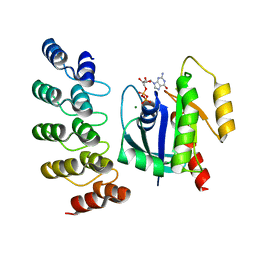 | | KRAS G13C complex with GDP imaged on a cryo-EM imaging scaffold | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Castells-Graells, R, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2023-02-09 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure determination of small therapeutic protein targets at 3 angstrom -resolution using a rigid imaging scaffold.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2RCF
 
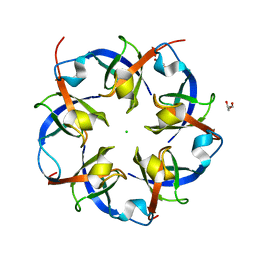 | | Carboxysome Shell protein, OrfA from H. Neapolitanus | | Descriptor: | CHLORIDE ION, GLYCEROL, Unidentified carboxysome polypeptide | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
2RIF
 
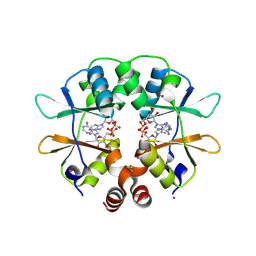 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CESIUM ION, Conserved protein with 2 CBS domains | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon.
J.Mol.Biol., 380, 2008
|
|
2RIH
 
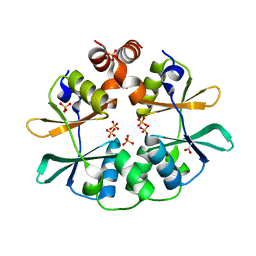 | | CBS domain protein PAE2072 from Pyrobaculum aerophilum | | Descriptor: | ACETIC ACID, Conserved protein with 2 CBS domains, SULFATE ION | | Authors: | Lee, T.M, King, N.P, Sawaya, M.R, Cascio, D, Yeates, T.O. | | Deposit date: | 2007-10-10 | | Release date: | 2008-06-17 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and Functional Implications of an AMP-Binding Cystathionine beta-Synthase Domain Protein from a Hyperthermophilic Archaeon
J.Mol.Biol., 380, 2008
|
|
1NOM
 
 | |
4IVJ
 
 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, I222 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-23 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (7.351 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4IQ4
 
 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P21212 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-10 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4ITV
 
 | | Structure of a 16 nm protein cage designed by fusing symmetric oligomeric domains, triple mutant, P212121 form | | Descriptor: | Non-haem bromoperoxidase BPO-A2, Matrix protein 1 | | Authors: | Lai, Y.-T, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2013-01-18 | | Release date: | 2013-07-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.598 Å) | | Cite: | Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
J.Am.Chem.Soc., 135, 2013
|
|
2Y5Y
 
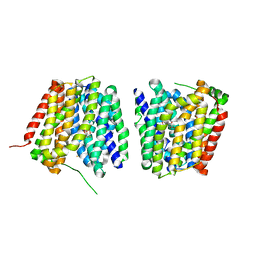 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y29
 
 | | Structure of segment KLVFFA from the amyloid-beta peptide (Ab, residues 16-21), alternate polymorph III | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Colletier, J, Laganowsky, A, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Basis for Amyloid-{Beta} Polymorphism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5E5V
 
 | | Structure of amyloid forming peptide NFGAILS (residues 22-28) from Islet Amyloid Polypeptide | | Descriptor: | NFGAILS (22-28) from islet amyloid polypeptide, synthesized | | Authors: | Soriaga, A.B, Macdonald, R, Sawaya, M.R, Sangwan, S, Eisenberg, D. | | Deposit date: | 2015-10-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal Structures of IAPP Amyloidogenic Segments Reveal a Novel Packing Motif of Out-of-Register Beta Sheets.
J.Phys.Chem.B, 120, 2016
|
|
5FOZ
 
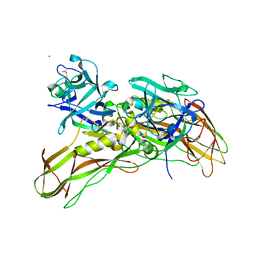 | | De novo structure of the binary mosquito larvicide BinAB at pH 10 | | Descriptor: | LARVICIDAL TOXIN PROTEIN, SODIUM ION, TOXIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5FOY
 
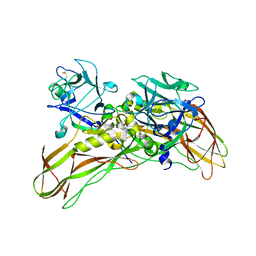 | | De novo structure of the binary mosquito larvicide BinAB at pH 7 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
5DRK
 
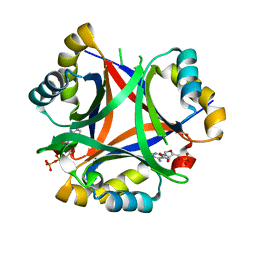 | | 2.3 Angstrom Structure of CPII, a nitrogen regulatory PII-like protein from Thiomonas intermedia K12, bound to ADP, AMP and bicarbonate. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Wheatley, N.M, Ngo, J, Cascio, D, Sawaya, M.R, Yeates, T.O. | | Deposit date: | 2015-09-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A PII-Like Protein Regulated by Bicarbonate: Structural and Biochemical Studies of the Carboxysome-Associated CPII Protein.
J.Mol.Biol., 428, 2016
|
|
