1OFT
 
 | |
1OFU
 
 | |
2W6D
 
 | | BACTERIAL DYNAMIN-LIKE PROTEIN LIPID TUBE BOUND | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, DYNAMIN FAMILY PROTEIN, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Low, H.H, Sachse, C, Amos, L.A, Lowe, J. | | Deposit date: | 2008-12-18 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of a Bacterial Dynamin-Like Protein Lipid Tube Provides a Mechanism for Assembly and Membrane Curving.
Cell(Cambridge,Mass.), 139, 2009
|
|
2XKB
 
 | |
2XKA
 
 | |
2VH1
 
 | | Crystal structure of bacterial cell division protein FtsQ from E.coli | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Naninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
2XIT
 
 | | Crystal structure of monomeric MipZ | | Descriptor: | MIPZ | | Authors: | Kiekebusch, D, Michie, K.A, Essen, L.O, Lowe, J, Thanbichler, M. | | Deposit date: | 2010-06-30 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Localized Dimerization and Nucleoid Binding Drive Gradient Formation by the Bacterial Cell Division Inhibitor Mipz.
Mol.Cell, 46, 2012
|
|
2VAM
 
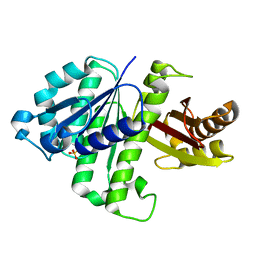 | | FtsZ B. subtilis | | Descriptor: | CELL DIVISION PROTEIN FTSZ, SULFATE ION | | Authors: | Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Conformational Variability of Ftsz
J.Mol.Biol., 373, 2007
|
|
2VAP
 
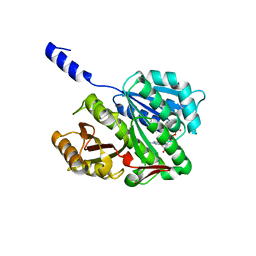 | | FtsZ GDP M. jannaschii | | Descriptor: | CELL DIVISION PROTEIN FTSZ HOMOLOG 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Conformational Variability of Ftsz
J.Mol.Biol., 373, 2007
|
|
2VH2
 
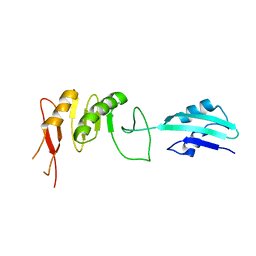 | | Crystal structure of cell divison protein FtsQ from Yersinia enterecolitica | | Descriptor: | CELL DIVISION PROTEIN FTSQ | | Authors: | van den Ent, F, Vinkenvleugel, T, Ind, A, West, P, Veprintsev, D, Nanninga, N, den Blaauwen, T, Lowe, J. | | Deposit date: | 2007-11-16 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and Mutational Analysis of Cell Division Protein Ftsq
Mol.Microbiol., 68, 2008
|
|
2XJ9
 
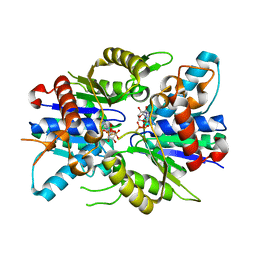 | | Dimer Structure of the bacterial cell division regulator MipZ | | Descriptor: | MAGNESIUM ION, MIPZ, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Michie, K.A, Lowe, J. | | Deposit date: | 2010-07-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Localized Dimerization and Nucleoid Binding Drive Gradient Formation by the Bacterial Cell Division Inhibitor Mipz.
Mol.Cell, 46, 2012
|
|
2VAW
 
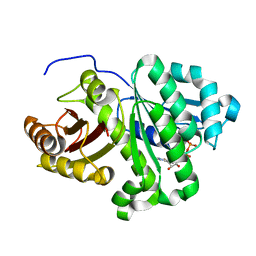 | | FtsZ Pseudomonas aeruginosa GDP | | Descriptor: | CELL DIVISION PROTEIN FTSZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights Into the Conformational Variability of Ftsz
J.Mol.Biol., 373, 2007
|
|
2XJ4
 
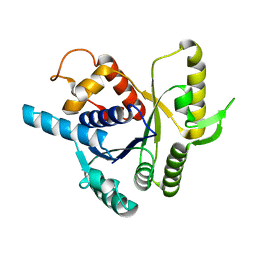 | |
1HYQ
 
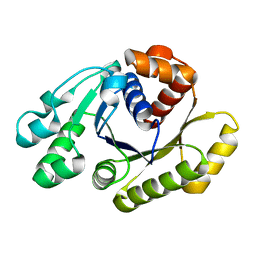 | |
1JCE
 
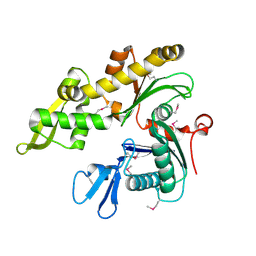 | |
1JCG
 
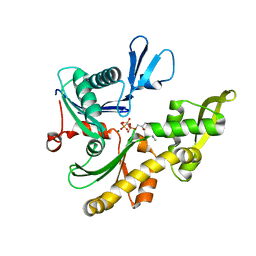 | | MREB FROM THERMOTOGA MARITIMA, AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ROD SHAPE-DETERMINING PROTEIN MREB | | Authors: | van den Ent, F, Amos, L.A, Lowe, J. | | Deposit date: | 2001-06-09 | | Release date: | 2001-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Prokaryotic origin of the actin cytoskeleton.
Nature, 413, 2001
|
|
1JCF
 
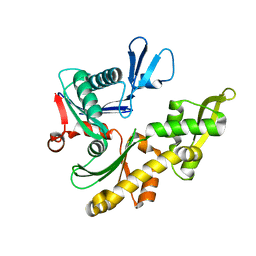 | |
1PMA
 
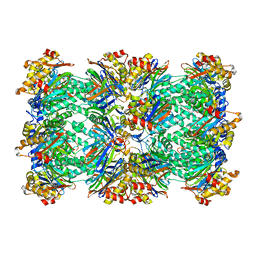 | | PROTEASOME FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PROTEASOME | | Authors: | Loewe, J, Stock, D, Jap, B, Zwickl, P, Baumeister, W, Huber, R. | | Deposit date: | 1994-12-19 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution.
Science, 268, 1995
|
|
3M6A
 
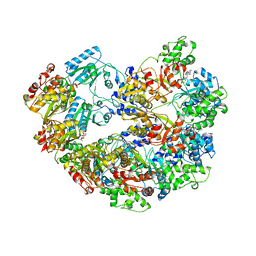 | |
7DG5
 
 | |
3M65
 
 | |
4K7P
 
 | |
8DKN
 
 | | PPARg bound to T0070907 and Co-R peptide | | Descriptor: | 2-chloro-5-nitro-N-(pyridin-4-yl)benzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1 peptide, ... | | Authors: | Larsen, N.A, Tsai, J. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DKV
 
 | | PPARg bound to JTP-426467 and Co-R peptide | | Descriptor: | 2-chloro-N-[4-(5-methyl-1,3-benzoxazol-2-yl)phenyl]-5-nitrobenzamide, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Nuclear receptor corepressor 1, ... | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
8DSZ
 
 | | PPARg bound to partial agonist H3B-487 | | Descriptor: | (2R)-2-{5-[(5-{[(1R)-1-(4-tert-butylphenyl)ethyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
