4ZRI
 
 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
1A2A
 
 | | AGKISTROTOXIN, A PHOSPHOLIPASE A2-TYPE PRESYNAPTIC NEUROTOXIN FROM AGKISTRODON HALYS PALLAS | | Descriptor: | CHLORIDE ION, PHOSPHOLIPASE A2 | | Authors: | Tang, L, Zhou, Y, Lin, Z. | | Deposit date: | 1997-12-25 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of agkistrodotoxin, a phospholipase A2-type presynaptic neurotoxin from agkistrodon halys pallas.
J.Mol.Biol., 282, 1998
|
|
1C1J
 
 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
5K7K
 
 | |
4LX4
 
 | | Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglucanase(Endo-1,4-beta-glucanase)protein | | Authors: | Dutoit, R, Delsaute, M, Berlemont, R, Van Elder, D, Galleni, M, Bauvois, C. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.556 Å) | | Cite: | Crystal structure determination of Pseudomonas stutzeri A1501 endoglucanase Cel5A: the search for a molecular basis for glycosynthesis in GH5_5 enzymes.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6UM8
 
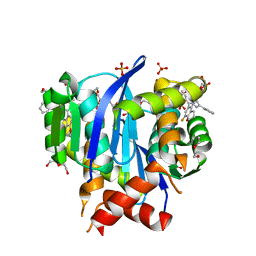 | | HIV Integrase in complex with Compound-14 | | Descriptor: | (2S)-tert-butoxy[7-(8-fluoro-5-methyl-3,4-dihydro-2H-1-benzopyran-6-yl)-5-methyl-2-phenylpyrazolo[1,5-a]pyrimidin-6-yl]acetic acid, DI(HYDROXYETHYL)ETHER, Integrase, ... | | Authors: | Khan, J.A, Kish, K. | | Deposit date: | 2019-10-09 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Discovery and Optimization of Novel Pyrazolopyrimidines as Potent and Orally Bioavailable Allosteric HIV-1 Integrase Inhibitors.
J.Med.Chem., 63, 2020
|
|
3T48
 
 | |
3T46
 
 | |
3T49
 
 | |
3T47
 
 | |
3HR5
 
 | |
3T4A
 
 | | Structure of a truncated form of Staphylococcal Complement Inhibitor B bound to human C3c at 3.4 Angstrom resolution | | Descriptor: | Complement C3 beta chain, Complement C3c alpha' chain fragment 1, Complement C3c alpha' chain fragment 2, ... | | Authors: | Garcia, B.L, Geisbrecht, B.V, Summers, B.J. | | Deposit date: | 2011-07-25 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Diversity in the C3b Convertase Contact Residues and Tertiary Structures of the Staphylococcal Complement Inhibitor (SCIN) Protein Family.
J.Biol.Chem., 287, 2012
|
|
6F6Z
 
 | | Mouse Thymidylate Synthase Cocrystallized with N(4)OHdCMP and Soaked in Methylenetetrahydrofolate | | Descriptor: | (2~{S})-2-[[4-[[(6~{R})-2-azanyl-4-oxidanylidene-5,6,7,8-tetrahydro-1~{H}-pteridin-6-yl]methyl-methyl-amino]phenyl]carbonylamino]pentanedioic acid, 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), Thymidylate synthase | | Authors: | Wilk, P, Jarmula, A, Maj, P, Rode, W. | | Deposit date: | 2017-12-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Molecular Mechanism of Thymidylate Synthase Inhibition by N4-Hydroxy-dCMP in View of Spectrophotometric and Crystallographic Studies
Int J Mol Sci, 22, 2021
|
|
5FOA
 
 | | Crystal Structure of Human Complement C3b in complex with DAF (CCP2-4) | | Descriptor: | COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA CHAIN, DECAY ACCELERATING FACTOR, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.188 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO9
 
 | | Crystal Structure of Human Complement C3b in Complex with CR1 (CCP15- 17) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO7
 
 | | Crystal Structure of Human Complement C3b at 2.8 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
1VKL
 
 | | RABBIT MUSCLE PHOSPHOGLUCOMUTASE | | Descriptor: | NICKEL (II) ION, PHOSPHOGLUCOMUTASE | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural changes at the metal ion binding site during the phosphoglucomutase reaction.
Biochemistry, 32, 1993
|
|
9J0C
 
 | |
8UAB
 
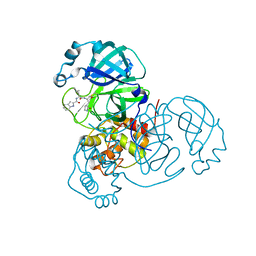 | | SARS-CoV-2 main protease (Mpro) complex with AC1115 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | DuPrez, K.T, Chao, A, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19
Med, 5, 2024
|
|
8UAC
 
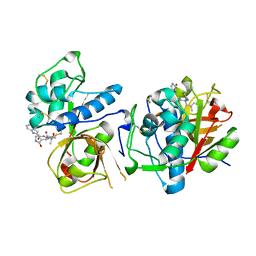 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19.
Med, 5, 2024
|
|
5FOB
 
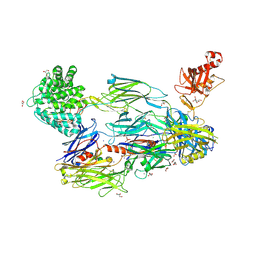 | | Crystal Structure of Human Complement C3b in complex with Smallpox Inhibitor of Complement (SPICE) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COMPLEMENT C3 BETA CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
5FO8
 
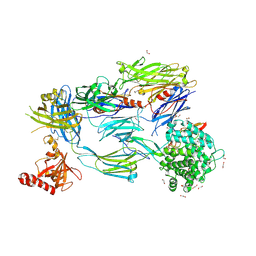 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
4O9B
 
 | | The Structure of CC1-IH in human STIM1. | | Descriptor: | CADMIUM ION, Stromal interaction molecule 1 | | Authors: | Cui, B, Yang, X, Li, S, Shen, Y. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1.
Plos One, 8, 2013
|
|
5V6G
 
 | | Crystal structure of Influenza A virus Matrix Protein M1(NLS-88R) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
5V7B
 
 | | Crystal structure of Influenza A virus matrix protein M1 (NLS-88E) | | Descriptor: | Matrix protein 1 | | Authors: | Musayev, F.N, Safo, M.K, Desai, U.R, Xie, H, Mosier, P.D, Chiang, M.-J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Maintaining pH-dependent conformational flexibility of M1 is critical for efficient influenza A virus replication.
Emerg Microbes Infect, 6, 2017
|
|
