8Z0O
 
 | | Cryo-EM structure of chitin synthase | | Descriptor: | MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, chitin synthase | | Authors: | Zhang, X, Niu, S, Li, P, Bi, Y. | | Deposit date: | 2024-04-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chitin Translocation Is Functionally Coupled with Synthesis in Chitin Synthase.
Int J Mol Sci, 25, 2024
|
|
4IDU
 
 | |
4JNG
 
 | | Schmallenberg virus nucleoprotein-RNA complex | | Descriptor: | Nucleocapsid protein, RNA (42-MER) | | Authors: | Dong, H.H, Li, P, Bottcher, B, Elliott, R.M, Dong, C.J. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of Schmallenberg orthobunyavirus nucleoprotein-RNA complex reveals a novel RNA sequestration mechanism.
Rna, 19, 2013
|
|
4L1L
 
 | | Rat PKC C2 domain bound to CD | | Descriptor: | CADMIUM ION, Protein kinase C alpha type, SULFATE ION | | Authors: | Morales, K.M, Yang, Y, Long, Z, Li, P, Taylor, A.B, Hart, P.J, Igumenova, T.I. | | Deposit date: | 2013-06-03 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cd(2+) as a ca(2+) surrogate in protein-membrane interactions: isostructural but not isofunctional.
J.Am.Chem.Soc., 135, 2013
|
|
7WQY
 
 | |
6ATV
 
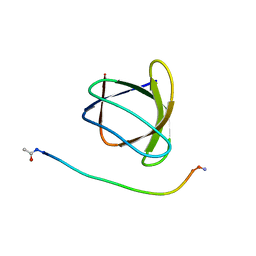 | | The molecular mechanisms by which NS1 of the 1918 Spanish influenza A virus hijack host protein-protein interactions | | Descriptor: | Adapter molecule crk, proline-rich motif in IAV-NS1 | | Authors: | Shen, Q, Zeng, D, Zhao, B, Li, P, Cho, J.H. | | Deposit date: | 2017-08-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular Mechanisms of Tight Binding through Fuzzy Interactions.
Biophys. J., 114, 2018
|
|
7W3S
 
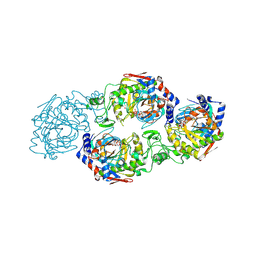 | | The complex structure of Larg1-ADPr from Legionella pneumophila | | Descriptor: | Type IV secretion protein Dot, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Ouyang, S, Guan, H, Li, P. | | Deposit date: | 2021-11-26 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Legionella pneumophila temporally regulates the activity of ADP/ATP translocases by reversible ADP-ribosylation.
mLife, 2022
|
|
8Z7X
 
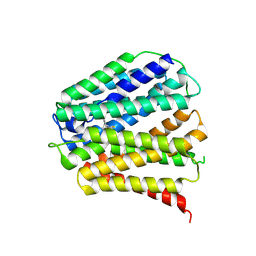 | | SLC19A3-Thiamine inward structure | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7R
 
 | | SLC19A3 apo structure | | Descriptor: | Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7T
 
 | | SLC19A3-Pyridoxine outward structure | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z80
 
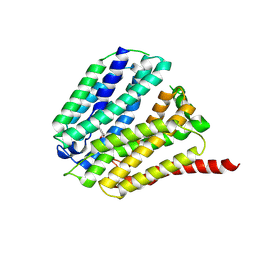 | | SLC19A2-Pyridoxine inward structure | | Descriptor: | 4,5-bis(hydroxymethyl)-2-methyl-pyridin-3-ol, Thiamine transporter 1 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7S
 
 | | SLC19A3-thiamine outward structure | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7W
 
 | | SLC19A3-Metformin outward structure | | Descriptor: | Metformin, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7V
 
 | | Slc19A3-Amprolium outward structure | | Descriptor: | Amprolium, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7Y
 
 | | SLC19A3-Fedratinib inward structure | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7U
 
 | | SLC19A3-Fedratinib outward structure | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Thiamine transporter 2 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
8Z7Z
 
 | | SLC19A2-Thiamine inward structure | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Thiamine transporter 1 | | Authors: | Li, P.P, Zhu, Z.N, Wang, Y, Gao, P, Qu, Q.H. | | Deposit date: | 2024-04-21 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Substrate transport and drug interaction of human thiamine transporters SLC19A2/A3.
Nat Commun, 15, 2024
|
|
5KEV
 
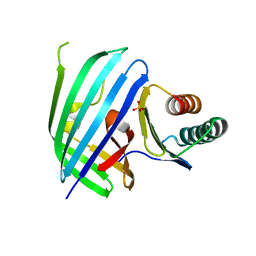 | |
5KEW
 
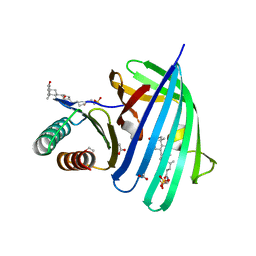 | | Vibrio parahaemolyticus VtrA/VtrC complex bound to the bile salt taurodeoxycholate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Tomchick, D.R, Orth, K, Rivera-Cancel, G. | | Deposit date: | 2016-06-10 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Bile salt receptor complex activates a pathogenic type III secretion system.
Elife, 5, 2016
|
|
5B25
 
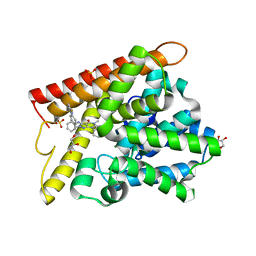 | | Crystal structure of human PDE1B with inhibitor 3 | | Descriptor: | (11R,15S)-4-{[4-(6-fluoropyridin-2-yl)phenyl]methyl}-8-methyl-5-(phenylamino)-1,3,4,8,10-pentaazatetracyclo[7.6.0.02,6.011,15]pentadeca-2,5,9-trien-7-one, Calcium/calmodulin-dependent 3',5'-cyclic nucleotide phosphodiesterase 1B, GLYCEROL, ... | | Authors: | Ida, K, Lane, W, Snell, G, Sogabe, S. | | Deposit date: | 2016-01-07 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Phosphodiesterase 1 for the Treatment of Cognitive Impairment Associated with Neurodegenerative and Neuropsychiatric Diseases
J.Med.Chem., 59, 2016
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5JAP
 
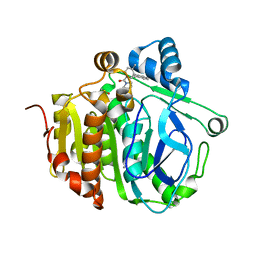 | |
9KQK
 
 | | Crystal structure of HSA in complex with AmpHecy | | Descriptor: | 2-[[6-[5-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]thiophen-2-yl]naphthalen-2-yl]-methyl-amino]ethanol, Albumin | | Authors: | Chen, X, Shi, M.W, Ge, Y.H, Fang, B, Li, L. | | Deposit date: | 2024-11-26 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Fluorogen-Activating Human Serum Albumin for Mitochondrial Nanoscale Imaging.
Adv Mater, 2025
|
|
1TW6
 
 | | Structure of an ML-IAP/XIAP chimera bound to a 9mer peptide derived from Smac | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Vucic, D, Wallweber, H.J.A, Das, K, Shin, H, Elliott, L.O, Kadkhodayan, S, Deshayes, K, Salvesen, G.S, Fairbrother, W.J. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.713 Å) | | Cite: | Engineering ML-IAP to produce an extraordinarily potent caspase 9 inhibitor: implications for Smac-dependent anti-apoptotic activity of ML-IAP
Biochem.J., 385, 2005
|
|
1KPR
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ALPHA CHAIN, ... | | Authors: | Holmes, M.A, Strong, R.K. | | Deposit date: | 2002-01-02 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | HLA-E allelic variants: Correlating differential expression, peptide affinities, crystal structures and thermal stabilities
J.Biol.Chem., 278, 2003
|
|
