8CW3
 
 | | 20us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWB
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWE
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVV
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWD
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVW
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWF
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
4U72
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (A260G mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4U74
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (G262A mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
4YB9
 
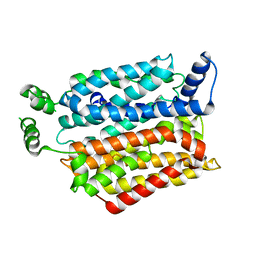 | | Crystal structure of the Bovine Fructose transporter GLUT5 in an open inward-facing conformation | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5 | | Authors: | Verdon, G, Kang, H.J, Iwata, S, Drew, D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5.
Nature, 526, 2015
|
|
4YBQ
 
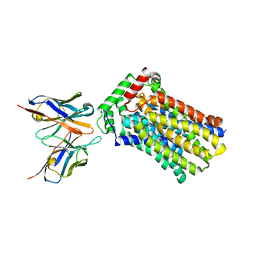 | | Rat GLUT5 with Fv in the outward-open form | | Descriptor: | Solute carrier family 2, facilitated glucose transporter member 5, antibody Fv fragment heavy chain, ... | | Authors: | Nomura, N, Shimamura, T, Iwata, S. | | Deposit date: | 2015-02-19 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structure and mechanism of the mammalian fructose transporter GLUT5
Nature, 526, 2015
|
|
7VWS
 
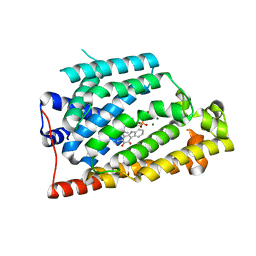 | | Carbazole Prenyl Transferase LvqB4 | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, LvqB4, MAGNESIUM ION, ... | | Authors: | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7VWT
 
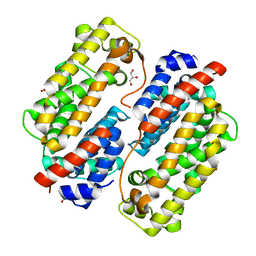 | | Carbazole Prenyl Transferase CqsB4 | | Descriptor: | 2-methyl-1-[(2R)-2-oxidanylpropyl]-9H-carbazole-3,4-dione, CqsB4, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suemune, H, Nagata, R, Kuzuyama, T, Nagano, S. | | Deposit date: | 2021-11-11 | | Release date: | 2022-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for the Prenylation Reaction of Carbazole-Containing Natural Products Catalyzed by Squalene Synthase-Like Enzymes.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7W8Y
 
 | | DMSPP- and Naplha-Me-Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-dimethylallyltryptophan synthase, ... | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8V
 
 | | DMSPP- and Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, SULFATE ION, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8X
 
 | | DMSPP- and 6-Me-Trp-bound dimethylallyl tryptophan synthase, IptA | | Descriptor: | (2S)-2-azanyl-3-(6-methyl-1H-indol-3-yl)propanoic acid, 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8W
 
 | | DMSPP- and 5-Me-Trp-bound 6-dimethylallyl tryptophan synthase, IptA | | Descriptor: | 5-methyl-L-tryptophan, 6-dimethylallyltryptophan synthase, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Suemune, H, Nagano, S, Tomoya, H. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
7W8U
 
 | | Crystal Structure of Indole Prenyltransferase IptA | | Descriptor: | 6-dimethylallyltryptophan synthase | | Authors: | Suemune, H, Nagano, S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of a 6-dimethylallyltryptophan synthase, IptA: Insights into substrate tolerance and enhancement of prenyltransferase activity.
Biochem.Biophys.Res.Commun., 593, 2022
|
|
1HQC
 
 | | STRUCTURE OF RUVB FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | ADENINE, MAGNESIUM ION, RUVB | | Authors: | Yamada, K, Kunishima, N, Mayanagi, K, Iwasaki, H, Morikawa, K. | | Deposit date: | 2000-12-15 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6LKM
 
 | | Crystal structure of Ribonucleotide reductase R1 subunit, RRM1 in complex with 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide | | Descriptor: | 5-chloro-N-((1S,2R)-2-(6-fluoro-2,3-dimethylphenyl)-1-(5-oxo-4,5-dihydro-1,3,4-oxadiazol-2-yl)propyl)-4-methyl-3,4-dihydro-2H-benzo[b][1,4]oxazine-8-sulfonamide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Miyahara, S, Chong, K.T, Suzuki, T. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | TAS1553, a novel small molecule ribonucleotide reductase (RNR) subunit interaction inhibitor, displays remarkable anti-tumor activity
To be published
|
|
8YFY
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 138, 2024
|
|
8YFZ
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
2D1J
 
 | | Factor Xa in complex with the inhibitor 2-[[4-[(5-chloroindol-2-yl)sulfonyl]piperazin-1-yl] carbonyl]thieno[3,2-b]pyridine n-oxide | | Descriptor: | 2-({4-[(5-CHLORO-1H-INDOL-2-YL)SULFONYL]PIPERAZIN-1-YL}CARBONYL)THIENO[3,2-B]PYRIDINE 4-OXIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological activity of non-basic compounds as factor Xa inhibitors: SAR study of S1 and aryl binding sites.
Bioorg.Med.Chem., 13, 2005
|
|
1WU1
 
 | |
7B7V
 
 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
