8B7P
 
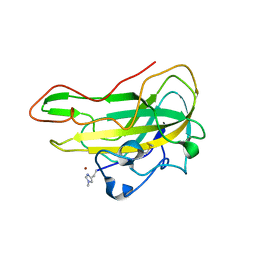 | | Crystal structure of an AA9 LPMO from Aspergillus nidulans, AnLPMOC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Endo-beta-1,4-glucanase D | | Authors: | Males, A, Rafael Fanchini Terrasan, C, Davies, G.J, Walton, P.H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterisation of lytic polysaccharide monooxygenases from Aspergillus nidulans
To Be Published
|
|
8BQA
 
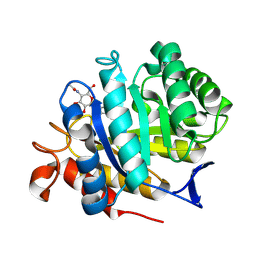 | | CjCel5B endo-glucanase bound to CB665 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, Endoglucanase, alpha-D-xylopyranose, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BQC
 
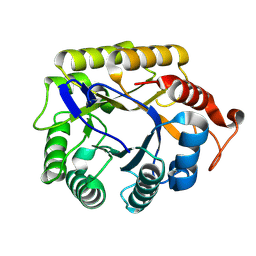 | | CjCel5B endo-glucanase | | Descriptor: | Endoglucanase | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BN7
 
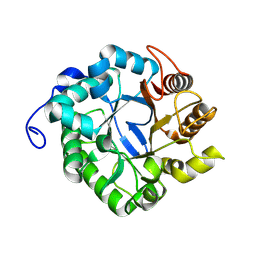 | | CjCel5C endo-glucanase | | Descriptor: | Cellulase, putative, cel5C | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2022-11-13 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BQB
 
 | | CjCel5C endo-glucanase bound to CB396 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, Cellulase, putative, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
8BC2
 
 | | Ligand-Free Structure of the decameric sulfofructose transaldolase BmSF-TAL | | Descriptor: | Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8BC4
 
 | | Cryo-EM Structure of a BmSF-TAL - Sulfofructose Schiff Base Complex in symmetry group C1 | | Descriptor: | (2~{R},3~{S},4~{S})-2,3,4,6-tetrakis(oxidanyl)hexane-1-sulfonic acid, Transaldolase | | Authors: | Snow, A.J.D, Sharma, M, Blaza, J, Davies, G.J. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and mechanism of sulfofructose transaldolase, a key enzyme in sulfoquinovose metabolism.
Structure, 31, 2023
|
|
8B0B
 
 | | Crystal structure of human heparanase in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8B0E
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB158 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-3,4,6-tris(oxidanyl)-2-[2-[2,2,2-tris(fluoranyl)ethanoylamino]ethoxy]cyclohexane-1-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8B0D
 
 | | Crystal structure of beta-glucuronidase from Acidobacterium capsulatum in complex with covalent inhibitor VB151 | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},6~{S})-2-(2-acetamidoethoxy)-3,4,6-tris(oxidanyl)cyclohexane-1-carboxylic acid, ALANINE, SULFATE ION, ... | | Authors: | Armstrong, Z, Davies, G.J. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | 4-O-Substituted Glucuronic Cyclophellitols are Selective Mechanism-Based Heparanase Inhibitors.
Chemmedchem, 18, 2023
|
|
8C4I
 
 | |
8CM9
 
 | | Structure of human O-GlcNAc transferase in complex with UDP and tP11 | | Descriptor: | PHE-MET-PRO-LYS-TYR-SER-ILE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2023-02-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9GCB
 
 | | DUF4198 protein from Ideonella sakaiensis with Ni bound | | Descriptor: | MALONATE ION, NICKEL (II) ION, Nickel ECF transporter, ... | | Authors: | Franco Cairo, J.P.L, Correa, T.L.R, Offen, W.A, Nairn, A, Walton, J, Davies, G.J, Walton, P.H, Sweeney, S.T. | | Deposit date: | 2024-08-01 | | Release date: | 2025-09-10 | | Last modified: | 2025-10-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Signal-strapping as a protein-sequence search method for the discovery of metalloproteins.
Nat Commun, 16, 2025
|
|
8OHT
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with competitive inhibitor derrived from siastatin B | | Descriptor: | (3~{S},4~{S},5~{S},6~{R})-4,5,6-tris(oxidanyl)piperidine-3-carboxylic acid, SULFATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OHU
 
 | | Crystal structure of Beta-glucuronidase from Acidobacterium capsulatum in complex with glucuronic acid configured isofagamine | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Armstrong, Z, Yurong, C, Wu, L, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-03-21 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
8OZ1
 
 | | CjCel5D endo-xyloglucanase bounc to CB665 covalent inhibitor | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, BORIC ACID, Cellulase, ... | | Authors: | McGregor, N.G.S, de Boer, C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A Multiplexing Activity-Based Protein-Profiling Platform for Dissection of a Native Bacterial Xyloglucan-Degrading System.
Acs Cent.Sci., 9, 2023
|
|
9HTA
 
 | | Dispersin from Terribacillus saccharophilus Dispts2 in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Dispersin | | Authors: | Males, A, Moroz, O.V, Blagova, E, Munch, A, Hansen, G.H, Johansen, A.H, Ostergaard, L.H, Segura, D.R, Eddenden, A, Due, A.V, Gudmand, M, Salomon, J, Sorensen, S.R, Franco Cairo, J.P.L, Pache, R.A, Vejborg, R.M, Bhosale, S, Vocadlo, D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Expansion of the diversity of dispersin scaffolds.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
8PEJ
 
 | | CjGH35 with a Galactosidase Activity-Based Probe | | Descriptor: | (1~{S},2~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, ACETATE ION, Beta-galactosidase, ... | | Authors: | Offen, W.A, Davies, G.J. | | Deposit date: | 2023-06-14 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The development of a broad-spectrum retaining beta-exo-galactosidase activity-based probe.
Org.Biomol.Chem., 21, 2023
|
|
9GWW
 
 | |
9GWU
 
 | |
9GWV
 
 | |
9GYY
 
 | |
9GYZ
 
 | |
8Q59
 
 | | Crystal structure of metal-dependent class II sulfofructose phosphate aldolase from Yersinia aldovae in complex with sulfofructose phosphate (YaSqiA-Zn-SFP) | | Descriptor: | (3~{S},4~{S})-2,3,4-tris(oxidanyl)-5-oxidanylidene-6-phosphonooxy-hexane-1-sulfonic acid, SODIUM ION, Tagatose-1,6-bisphosphate aldolase kbaY, ... | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
8Q5A
 
 | | Crystal structure of metal-dependent class II sulfofructosephosphate aldolase from Hafnia paralvei HpSqiA-Zn in complex with dihydroxyacetone phosphate (DHAP) | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Ketose-bisphosphate aldolase, ZINC ION | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2023-08-08 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defining the molecular architecture, metal dependence, and distribution of metal-dependent class II sulfofructose-1-phosphate aldolases.
J.Biol.Chem., 299, 2023
|
|
