2XKH
 
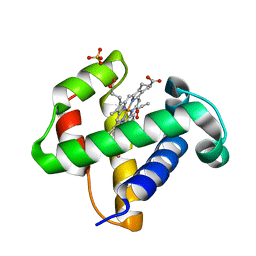 | | Xe derivative of C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XKI
 
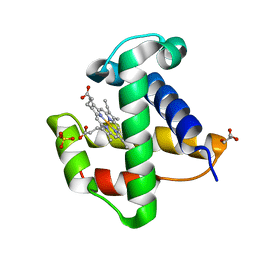 | | Aquo-met structure of C.lacteus mini-Hb | | Descriptor: | GLYCEROL, NEURAL HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
2XHZ
 
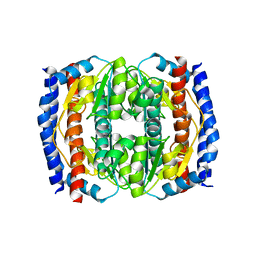 | | Probing the active site of the sugar isomerase domain from E. coli arabinose-5-phosphate isomerase via X-ray crystallography | | Descriptor: | ARABINOSE 5-PHOSPHATE ISOMERASE | | Authors: | Gourlay, L.J, Sommaruga, S, Nardini, M, Sperandeo, P, Deho, G, Polissi, A, Bolognesi, M. | | Deposit date: | 2010-06-24 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing the Active Site of the Sugar Isomerase Domain from E. Coli Arabinose-5-Phosphate Isomerase Via X-Ray Crystallography.
Protein Sci., 19, 2010
|
|
1HKU
 
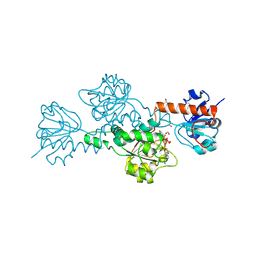 | | CtBP/BARS: a dual-function protein involved in transcription corepression and Golgi membrane fission | | Descriptor: | C-TERMINAL BINDING PROTEIN 3, FORMIC ACID, GLYCEROL, ... | | Authors: | Nardini, M, Spano, S, Cericola, C, Pesce, A, Massaro, A, Millo, E, Luini, A, Corda, D, Bolognesi, M. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-19 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ctbp/Bars: A Dual-Function Protein Involved in Transcription Co-Repression and Golgi Membrane Fission
Embo J., 22, 2003
|
|
2XKG
 
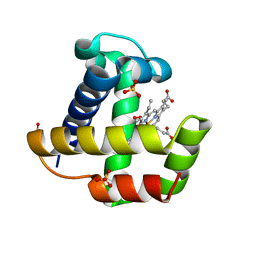 | | C.lacteus mini-Hb Leu86Ala mutant | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Dewilde, S, Capece, L, Marti, M.A, Congia, S, Salter, M.D, Blouin, G.C, Estrin, D.A, Ascenzi, P, Moens, L, Bolognesi, M, Olson, J.S. | | Deposit date: | 2010-07-08 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ligand Migration in the Apolar Tunnel of Cerebratulus Lacteus Mini-Hemoglobin.
J.Biol.Chem., 286, 2011
|
|
1S56
 
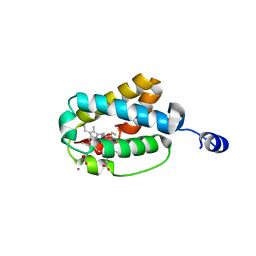 | | Crystal Structure of "Truncated" Hemoglobin N (HbN) from Mycobacterium tuberculosis, Soaked with Xe Atoms | | Descriptor: | CYANIDE ION, HEME C, Hemoglobin-like protein HbN, ... | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-20 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Heme-ligand tunneling in group I truncated hemoglobins
J.Biol.Chem., 279, 2004
|
|
1S61
 
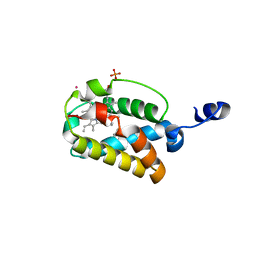 | | Crystal Structure of "Truncated" Hemoglobin N (HbN) from Mycobacterium tuberculosis, Soaked with Butyl-isocyanide | | Descriptor: | CYANIDE ION, Hemoglobin-like protein HbN, N-BUTYL ISOCYANIDE, ... | | Authors: | Milani, M, Pesce, A, Ouellet, Y, Dewilde, S, Friedman, J, Ascenzi, P, Guertin, M, Bolognesi, M. | | Deposit date: | 2004-01-22 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Heme-ligand tunneling in group I truncated hemoglobins
J.Biol.Chem., 279, 2004
|
|
3U10
 
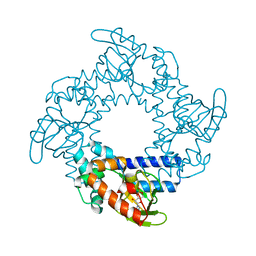 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
3U0Z
 
 | | Tetramerization dynamics of the C-terminus underlies isoform-specific cAMP-gating in HCN channels | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Lolicato, M, Nardini, M, Gazzarrini, S, Moller, S, Bertinetti, D, Herberg, F.W, Bolognesi, M, Martin, H, Fasolini, M, Bertrand, J.A, Arrigoni, C, Thiel, G, Moroni, A. | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Tetramerization dynamics of C-terminal domain underlies isoform-specific cAMP gating in hyperpolarization-activated cyclic nucleotide-gated channels.
J.Biol.Chem., 286, 2011
|
|
2VG6
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG5
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-chlorophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2VG7
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-iodophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
3UR0
 
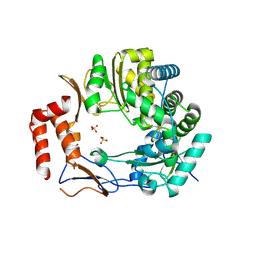 | | Crystal structures of murine norovirus RNA-dependent RNA polymerase in complex with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-02 | | Last modified: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
4RMS
 
 | |
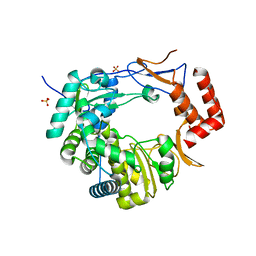
3UQS
 
 | |
4RMT
 
 | | Crystal structure of the D98N Beta-2 Microglobulin mutant | | Descriptor: | ACETATE ION, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | de Rosa, M, Bolognesi, M, Ricagno, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | Decoding the Structural Bases of D76N 2-Microglobulin High Amyloidogenicity through Crystallography and Asn-Scan Mutagenesis.
Plos One, 10, 2015
|
|
6R3W
 
 | | M.tuberculosis nitrobindin with a water molecule coordinated to the heme iron atom | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UPF0678 fatty acid-binding protein-like protein ERS007657_00996 | | Authors: | De Simone, G, di Masi, A, Polticelli, F, Pesce, A, Nardini, M, Bolognesi, M, Ciaccio, C, Coletta, M, Turilli, E.S, Fasano, M, Tognaccini, L, Smulevich, G, Abbruzzetti, S, Viappiani, C, Bruno, S, Ascenzi, P. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Mycobacterial and Human Nitrobindins: Structure and Function.
Antioxid.Redox Signal., 33, 2020
|
|
6R3Y
 
 | | M.tuberculosis nitrobindin with a cyanide molecule coordinated to the heme iron atom | | Descriptor: | CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE, UPF0678 fatty acid-binding protein-like protein ERS007657_00996 | | Authors: | De Simone, G, di Masi, A, Polticelli, F, Pesce, A, Nardini, M, Bolognesi, M, Ciaccio, C, Coletta, M, Turilli, E.S, Fasano, M, Tognaccini, L, Smulevich, G, Abbruzzetti, S, Viappiani, C, Bruno, S, Ascenzi, P. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mycobacterial and Human Nitrobindins: Structure and Function.
Antioxid.Redox Signal., 33, 2020
|
|
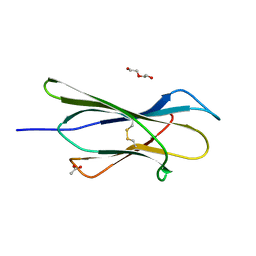
4RMR
 
 | |
4RMQ
 
 | |
3UPF
 
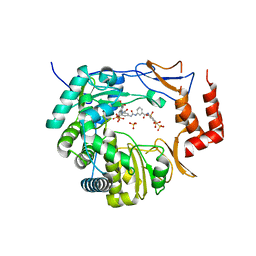 | | Crystal structure of murine norovirus RNA-dependent RNA polymerase bound to NF023 | | Descriptor: | 8-({3-[({3-[(4,6,8-trisulfonaphthalen-1-yl)carbamoyl]phenyl}carbamoyl)amino]benzoyl}amino)naphthalene-1,3,5-trisulfonic acid, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Milani, M, Mastrangelo, E, Bolognesi, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Inhibition of Norovirus RNA-Dependent RNA Polymerases.
J.Mol.Biol., 419, 2012
|
|
3TM6
 
 | |
4UII
 
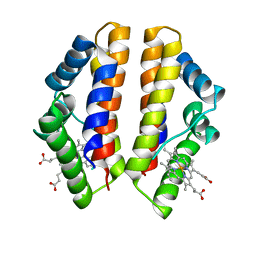 | | Crystal structure of the Azotobacter vinelandii globin-coupled oxygen sensor in the aquo-met form | | Descriptor: | GGDEF DOMAIN PROTEIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Germani, F, De Schutter, A, Pesce, A, Berghmans, H, Van Hauwaert, M.-L, Cuypers, B, Bruno, S, Mozzarelli, A, Moens, L, Van Doorslaer, S, Bolognesi, M, Nardini, M, Dewilde, S. | | Deposit date: | 2015-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.827 Å) | | Cite: | Azotobacter Vinelandii Globin-Coupled Oxygen Sensor is a Diguanylate Cyclase with a Biphasic Oxygen Dissociation
To be Published
|
|
4UIQ
 
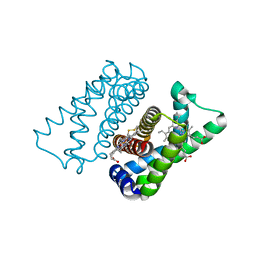 | | Isolated globin domain of the Bordetella pertussis globin-coupled sensor with a heme at the dimer interface | | Descriptor: | GLOBIN-COUPLED SENSOR WITH DIGUANYLATE CYCLASE ACTIVITY, GLYCEROL, IMIDAZOLE, ... | | Authors: | Germani, F, Donne, J, De Schutter, A, Pesce, A, Berghmans, H, Van Hauwaert, M.-L, Moens, L, Van Doorslaer, S, Bolognesi, M, Nardini, M, Dewilde, S. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of a Putative Allosteric Site in the Sensor Domain of Bordetella Pertussis Globin- Coupled Sensor for the Control of Biofilm Formation
To be Published
|
|
2XTL
 
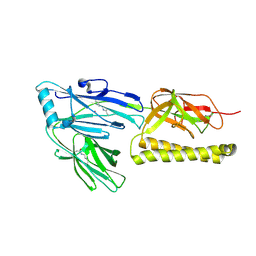 | | Structure of the major pilus backbone protein from Streptococcus Agalactiae | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, POTASSIUM ION | | Authors: | Rinauda, D, Gourlay, L.J, Soriano, M, Grandi, G, Bolognesi, M. | | Deposit date: | 2010-10-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Approach to Rationally Design a Chimeric Protein for an Effective Vaccine Against Group B Streptococcus Infections.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
