5LS8
 
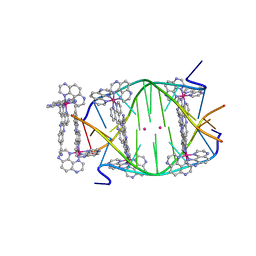 | | Light-activated ruthenium complex bound to a DNA quadruplex | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*TP*A)-3'), POTASSIUM ION, delta-[Ru(tap2-dppz-CN)]2+, ... | | Authors: | McQuaid, K.T, Abell, H, Hall, J.P, Cardin, C.J. | | Deposit date: | 2016-08-22 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Studies Reveal Enantiospecific Recognition of a DNA G-Quadruplex by a Ruthenium Polypyridyl Complex.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
1BQ5
 
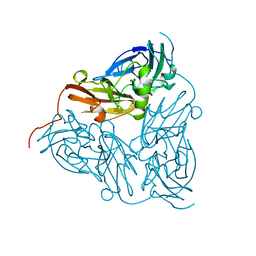 | | NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS GIFU 1051 | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Inoue, T, Gotowda, M, Deligeer, Suzuki, S, Kataoka, K, Yamaguchi, K, Watanabe, H, Goho, M, Yasushi, K.A.I. | | Deposit date: | 1998-08-21 | | Release date: | 1999-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Type 1 Cu structure of blue nitrite reductase from Alcaligenes xylosoxidans GIFU 1051 at 2.05 A resolution: comparison of blue and green nitrite reductases.
J.Biochem.(Tokyo), 124, 1998
|
|
3WIR
 
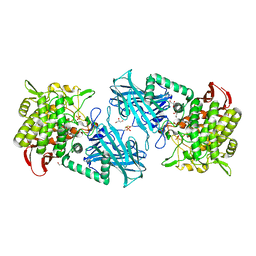 | | Crystal structure of kojibiose phosphorylase complexed with glucose | | Descriptor: | GLYCEROL, Kojibiose phosphorylase, PHOSPHATE ION, ... | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
1G04
 
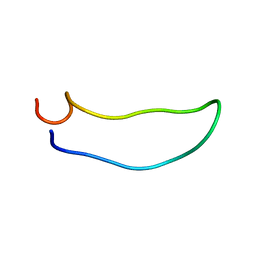 | | SOLUTION STRUCTURE OF SYNTHETIC 26-MER PEPTIDE CONTAINING 145-169 SHEEP PRION PROTEIN SEGMENT AND C-TERMINAL CYSTEINE | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Kozin, S.A, Bertho, G, Mazur, A.K, Rabesona, H, Girault, J.-P, Haertle, T, Takahashi, M, Debey, P, Hui Bon Hoa, G. | | Deposit date: | 2000-10-05 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sheep prion protein synthetic peptide spanning helix 1 and beta-strand 2 (residues 142-166) shows beta-hairpin structure in solution.
J.Biol.Chem., 276, 2001
|
|
3WIQ
 
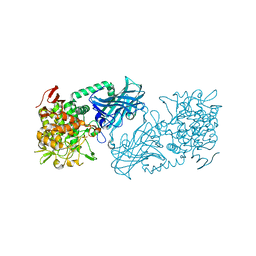 | | Crystal structure of kojibiose phosphorylase complexed with kojibiose | | Descriptor: | Kojibiose phosphorylase, SULFATE ION, alpha-D-glucopyranose-(1-2)-beta-D-glucopyranose | | Authors: | Okada, S, Yamamoto, T, Watanabe, H, Nishimoto, T, Chaen, H, Fukuda, S, Wakagi, T, Fushinobu, S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and mutational analysis of substrate recognition in kojibiose phosphorylase
Febs J., 281, 2014
|
|
1KSD
 
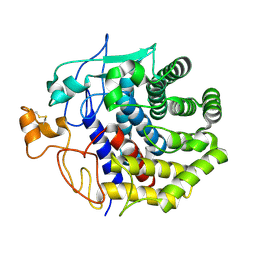 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 6.5. | | Descriptor: | CALCIUM ION, Endo-b-1,4-glucanase | | Authors: | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | Deposit date: | 2002-01-12 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KSC
 
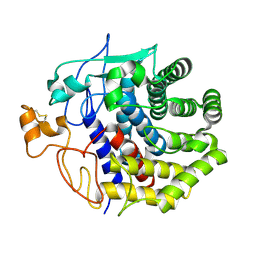 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 5.6. | | Descriptor: | CALCIUM ION, Endo-b-1,4-glucanase | | Authors: | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | Deposit date: | 2002-01-11 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KS8
 
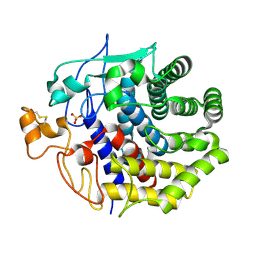 | | The structure of Endoglucanase from termite, Nasutitermes takasagoensis, at pH 2.5. | | Descriptor: | Endo-b-1,4-glucanase, SULFATE ION | | Authors: | Khademi, S, Guarino, L.A, Watanabe, H, Tokuda, G, Meyer, E.F. | | Deposit date: | 2002-01-11 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of an endoglucanase from termite, Nasutitermes takasagoensis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1S4T
 
 | | Solution structure of synthetic 21mer peptide spanning region 135-155 (in human numbering) of sheep prion protein | | Descriptor: | Major prion protein | | Authors: | Kozin, S.A, Lepage, C, Hui Bon Hoa, G, Rabesona, H, Mazur, A.K, Blond, A, Cheminant, M, Haertle, T, Debey, P, Rebuffat, S. | | Deposit date: | 2004-01-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Specific recognition between surface loop 2 (132-143) and helix 1 (144-154)
within sheep prion protein from in vitro studies of synthetic peptides
To be Published
|
|
1TV0
 
 | | Solution structure of cryptdin-4, the most potent alpha-defensin from mouse Paneth cells | | Descriptor: | Cryptdin-4 | | Authors: | Jing, W, Hunter, H.N, Tanabe, H, Ouellette, A.J, Vogel, H.J. | | Deposit date: | 2004-06-25 | | Release date: | 2005-01-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cryptdin-4, a Mouse Paneth Cell alpha-Defensin.
Biochemistry, 43, 2004
|
|
8IFJ
 
 | |
3QWM
 
 | | Crystal Structure of GEP100, the plextrin homology domain of IQ motif and SEC7 domain-containing protein 1 isoform a | | Descriptor: | IQ motif and SEC7 domain-containing protein 1 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Feller, S, Janning, M, Sabe, H, Krojer, T, Chaikuad, A, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-28 | | Release date: | 2011-04-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal Structure of GEP100, the plextrin homology domain of IQ motif and SEC7 domain-containing protein 1 isoform a
to be published
|
|
3APX
 
 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and chlorpromazine complex | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, ACETIC ACID, Alpha-1-acid glycoprotein 2 | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APW
 
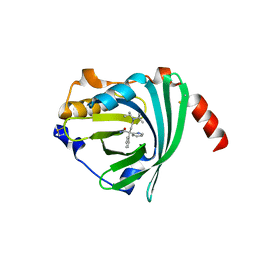 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and disopyramide complex | | Descriptor: | Alpha-1-acid glycoprotein 2, Disopyramide | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APU
 
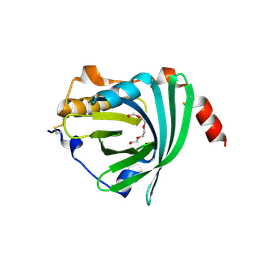 | | Crystal structure of the A variant of human alpha1-acid glycoprotein | | Descriptor: | Alpha-1-acid glycoprotein 2, TETRAETHYLENE GLYCOL | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
3APV
 
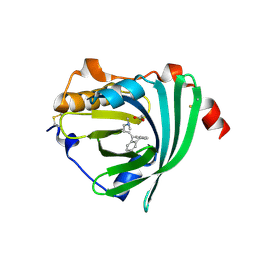 | | Crystal structure of the A variant of human alpha1-acid glycoprotein and amitriptyline complex | | Descriptor: | ACETIC ACID, Alpha-1-acid glycoprotein 2, Amitriptyline | | Authors: | Nishi, K, Ono, T, Nakamura, T, Fukunaga, N, Izumi, M, Watanabe, H, Suenaga, A, Maruyama, T, Yamagata, Y, Curry, S, Otagiri, M. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into differences in drug-binding selectivity between two forms of human alpha1-acid glycoprotein genetic variants, the A and F1*S forms.
J. Biol. Chem., 286, 2011
|
|
2DKC
 
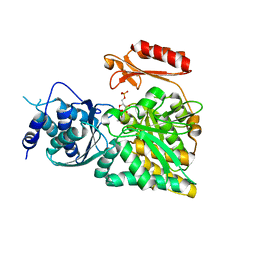 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the substrate complex | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DKD
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the product complex | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-galactopyranose, PHOSPHATE ION, Phosphoacetylglucosamine mutase, ... | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
2DKA
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
3AG1
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 280 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AG3
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ASE
 
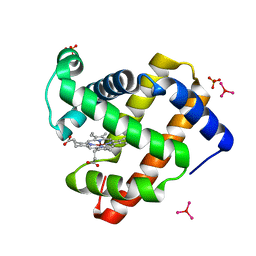 | | Crystal Structure of Zinc myoglobin soaked with Ru3O cluster | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING ZN, SULFATE ION, ... | | Authors: | Koshiyama, T, Shirai, M, Hikage, T, Tabe, H, Tanaka, K, Kitagawa, S, Ueno, T. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Post-Crystal Engineering of Zinc-Substituted Myoglobin to Construct a Long-Lived Photoinduced Charge-Separation System
Angew.Chem.Int.Ed.Engl., 2011
|
|
3AG2
 
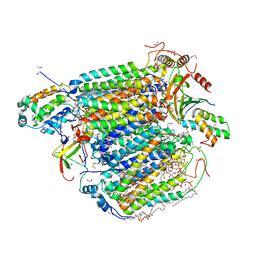 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AG4
 
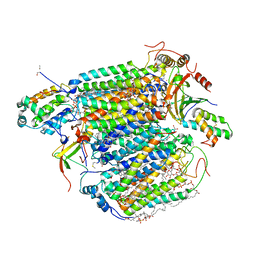 | | Bovine Heart Cytochrome c Oxidase in the Cyanide Ion-bound Fully Reduced State at 100 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2D1X
 
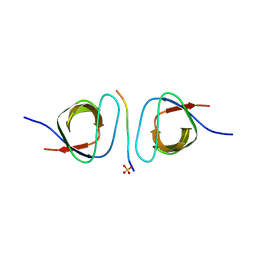 | | The crystal structure of the cortactin-SH3 domain and AMAP1-peptide complex | | Descriptor: | SULFATE ION, cortactin isoform a, proline rich region from development and differentiation enhancing factor 1 | | Authors: | Hashimoto, S, Hirose, M, Hashimoto, A, Morishige, M, Yamada, A, Hosaka, H, Akagi, K, Ogawa, E, Oneyama, C, Agatsuma, T, Okada, M, Kobayashi, H, Wada, H, Nakano, H, Ikegami, T, Nakagawa, A, Sabe, H. | | Deposit date: | 2005-09-01 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting AMAP1 and cortactin binding bearing an atypical src homology 3/proline interface for prevention of breast cancer invasion and metastasis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
