6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
4PCN
 
 | | Phosphotriesterase variant R22 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R22, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-04-16 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat. Chem. Biol., 12, 2016
|
|
6GW9
 
 | | Concanavalin A structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | CALCIUM ION, Concanavalin V, MAGNESIUM ION | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
6GWA
 
 | | Concanavalin B structure determined with data from the EuXFEL, the first MHz free electron laser | | Descriptor: | Concanavalin B | | Authors: | Gruenbein, M.L, Gorel, A, Stricker, M, Bean, R, Bielecki, J, Doerner, K, Hartmann, E, Hilpert, M, Kloos, M, Letrun, R, Sztuk-Dambietz, J, Mancuso, A, Meserschmidt, M, Nass-Kovacs, G, Ramilli, M, Roome, C.M, Sato, T, Doak, R.B, Shoeman, R.L, Foucar, L, Colletier, J.P, Barends, T.R.M, Stan, C, Schlichting, I. | | Deposit date: | 2018-06-22 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz data collection from protein microcrystals at an X-ray free-electron laser.
Nat Commun, 9, 2018
|
|
5MJD
 
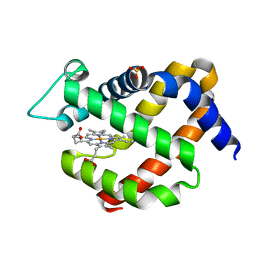 | | metNgb under oxygen at 80 bar | | Descriptor: | Neuroglobin, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5MJC
 
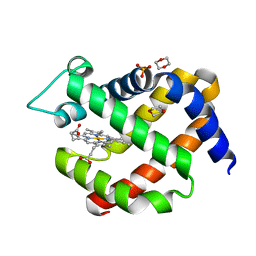 | | metNeuroglobin under oxygen at 50 bar | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, OXYGEN MOLECULE, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P, Vallone, B. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Iucrj, 6, 2019
|
|
5IKX
 
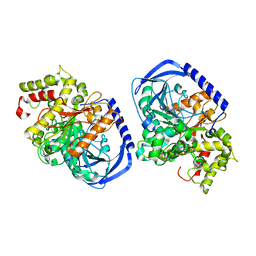 | |
4D65
 
 | |
4D64
 
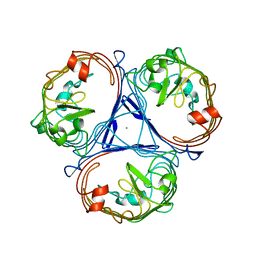 | |
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX4
 
 | | mosquitocidal Cry11Aa determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX6
 
 | | mosquitocidal Cry11Aa-E583Q determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX5
 
 | | mosquitocidal Cry11Aa-Y449F determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7QX7
 
 | | mosquitocidal Cry11Aa-F17Y determined at pH 7 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Aa | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P. | | Deposit date: | 2022-01-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
7R1E
 
 | | Mosquitocidal Cry11Ba determined at pH 10.4 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | GLYCEROL, Pesticidal crystal protein Cry11Ba | | Authors: | Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-02-02 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
5CH3
 
 | | E3 alpha-esterase-7 carboxylesterase | | Descriptor: | Carboxylic ester hydrolase | | Authors: | Correy, G, Mabbitt, P, Jackson, C.J. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography.
Structure, 24, 2016
|
|
5CH5
 
 | | E3 alpha-esterase-7 carboxylesterase | | Descriptor: | Carboxylic ester hydrolase, DIETHYL HYDROGEN PHOSPHATE | | Authors: | Correy, G, Mabbitt, P, Jackson, C.J. | | Deposit date: | 2015-07-10 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography.
Structure, 24, 2016
|
|
4XAY
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R8, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAZ
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphotriesterase variant PTE-R18, ZINC ION | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-16 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAG
 
 | | Cycles of destabilization and repair underlie the evolution of new enzyme function | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R6, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD3
 
 | | Phosphotriesterase variant E3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD6
 
 | | Phosphotriesterase Variant E2a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-E2, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XAF
 
 | | Cycles of destabilization and repair underlie evolutionary transitions in enzymes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R1, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-14 | | Release date: | 2015-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
4XD5
 
 | | Phosphotriesterase variant R2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R2, ... | | Authors: | Campbell, E, Kaltenbach, M, Tokuriki, N, Jackson, C.J. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
