3WAJ
 
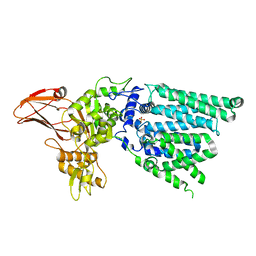 | |
2P0I
 
 | | Crystal structure of L-rhamnonate dehydratase from Gibberella zeae | | Descriptor: | GLYCEROL, L-rhamnonate dehydratase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Dickey, M, Logan, C, Gheyi, T, Wasserman, S.R, Smith, D, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of L-Rhamnonate Dehydratase from Gibberella Zeae
To be Published
|
|
3E2V
 
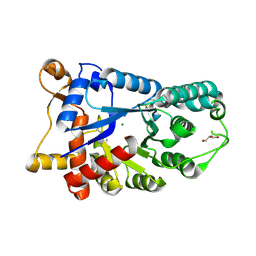 | | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae | | Descriptor: | 3'-5'-exonuclease, GLYCEROL, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Hu, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized amidohydrolase from Saccharomyces cerevisiae
To be Published
|
|
1GJO
 
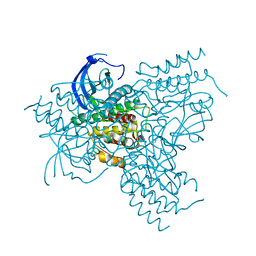 | | The FGFr2 tyrosine kinase domain | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, SULFATE ION | | Authors: | Ceska, T.A, Owens, R, Doyle, C, Hamlyn, P, Crabbe, T, Moffat, D, Davis, J, Martin, R, Perry, M.J. | | Deposit date: | 2001-07-31 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Fgfr2 Tyrosine Kinase Domain
To be Published
|
|
3WEV
 
 | | Crystal structure of the Schiff base intermediate of L-Lys epsilon-oxidase from Marinomonas mediterranea with L-Lys | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | Authors: | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | Deposit date: | 2013-07-12 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
3WH2
 
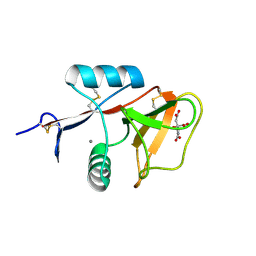 | | Human Mincle in complex with citrate | | Descriptor: | C-type lectin domain family 4 member E, CALCIUM ION, CITRATE ANION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-21 | | Release date: | 2013-10-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WHD
 
 | | C-type lectin, human MCL | | Descriptor: | C-type lectin domain family 4 member D, CALCIUM ION | | Authors: | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | Deposit date: | 2013-08-24 | | Release date: | 2013-10-23 | | Last modified: | 2013-12-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3H75
 
 | | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens | | Descriptor: | GLYCEROL, Periplasmic sugar-binding domain protein, SULFATE ION | | Authors: | Kumaran, D, Mahmood, A, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a Periplasmic Sugar-binding protein from the Pseudomonas fluorescens
To be Published
|
|
3H7D
 
 | | The crystal structure of the cathepsin K Variant M5 in complex with chondroitin-4-sulfate | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Cherney, M.M, Kienetz, M, Bromme, D, James, M.N.G. | | Deposit date: | 2009-04-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions.
J.Biol.Chem., 286, 2011
|
|
3ED3
 
 | | Crystal Structure of the Yeast Dithiol/Disulfide Oxidoreductase Mpd1p | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein disulfide-isomerase MPD1 | | Authors: | Vitu, E, Greenblatt, H.M, Fass, D. | | Deposit date: | 2008-09-02 | | Release date: | 2008-11-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Yeast Mpd1p reveals the structural diversity of the protein disulfide isomerase family
J.Mol.Biol., 384, 2008
|
|
1FVV
 
 | | THE STRUCTURE OF CDK2/CYCLIN A IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(7-OXO-7H-THIAZOLO[5,4-E]INDOL-8-YLMETHYL)-AMINO]-N-PYRIDIN-2-YL-BENZENESULFONAMIDE, CYCLIN A, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1GMN
 
 | | CRYSTAL STRUCTURES OF NK1-HEPARIN COMPLEXES REVEAL THE BASIS FOR NK1 ACTIVITY AND ENABLE ENGINEERING OF POTENT AGONISTS OF THE MET RECEPTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEPATOCYTE GROWTH FACTOR | | Authors: | Lietha, D, Chirgadze, D.Y, Mulloy, B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2001-09-19 | | Release date: | 2001-10-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Nk1-Heparin Complexes Reveal the Basis for Nk1 Activity and Enable Engineering of Potent Agonists of the met Receptor
Embo J., 20, 2001
|
|
3GTO
 
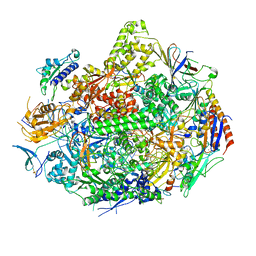 | | Backtracked RNA polymerase II complex with 15mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3HC0
 
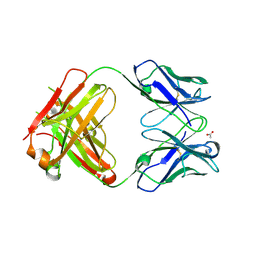 | | BHA10 IgG1 wild-type Fab - antibody directed at human LTBR | | Descriptor: | ACETATE ION, IMMUNOGLOBULIN IGG1 FAB, HEAVY CHAIN, ... | | Authors: | Arndt, J.W, Jordan, J.L, Lugovskoy, A, Wang, D. | | Deposit date: | 2009-05-05 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural understanding of stabilization patterns in engineered bispecific Ig-like antibody molecules
Proteins, 77, 2009
|
|
3EIV
 
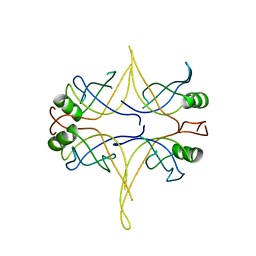 | |
3HCL
 
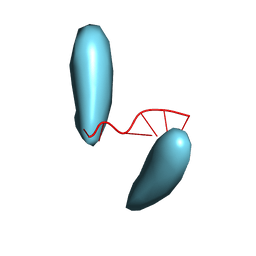 | | Helical superstructures in a DNA oligonucleotide crystal | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*AP*T)-3') | | Authors: | De Luchi, D, Martinez de Ilarduya, I, Subirana, J.A, Uson, I, Campos, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A geometric approach to the crystallographic solution of nonconventional DNA structures: helical superstructures of d(CGATAT)
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3EKB
 
 | |
2P0A
 
 | | The crystal structure of human synapsin III (SYN3) in complex with AMPPNP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Turnbull, A.P, Phillips, C, Pike, A.C.W, Elkins, J.M, Gileadi, C, Salah, E, Niesen, F.H, Burgess, N, Gileadi, O, Gorrec, F, Umeano, C, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C.H, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of human synapsin III (SYN3) in complex with AMPPNP
To be Published
|
|
1G0O
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND PYROQUILON | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYROQUILON, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Liao, D, Basarab, G.S, Gatenby, A.A, Valent, B, Jordan, D.B. | | Deposit date: | 2000-10-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of trihydroxynaphthalene reductase-fungicide complexes: implications for structure-based design and catalysis.
Structure, 9, 2001
|
|
1GKN
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
1G1F
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A TRI-PHOSPHORYLATED PEPTIDE (RDI(PTR)ETD(PTR)(PTR)RK) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE 1B, TRI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1GN0
 
 | | Escherichia coli GlpE sulfurtransferase soaked with KCN | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, THIOSULFATE SULFURTRANSFERASE GLPE | | Authors: | Spallarossa, A, Donahue, J.T, Larson, T.J, Bolognesi, M, Bordo, D. | | Deposit date: | 2001-10-01 | | Release date: | 2001-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Escherichia Coli Glpe is a Prototype Sulfurtransferase for the Single-Domain Rhodanese Homology Superfamily
Structure, 9, 2001
|
|
4A8N
 
 | | Protein crystallization and microgravity: glucose isomerase crystals grown during the PCDF-PROTEIN mission | | Descriptor: | COBALT (II) ION, GLYCEROL, XYLOSE ISOMERASE | | Authors: | Decanniere, K, Patino-Lopez, L.-D, Sleutel, M, Evrard, C, Van De Weerdt, C, Haumont, E, Gavira, J.A, Otalora, F, Maes, D. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Crystallization and Microgravity: Glucose Isomerase Crystals Grown During the Pcdf-Protein Mission
To be Published
|
|
3H4P
 
 | | Proteasome 20S core particle from Methanocaldococcus jannaschii | | Descriptor: | Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Jeffrey, P.D, Zhang, F, Hu, M, Tian, G, Zhang, P, Finley, D, Shi, Y. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural Insights into the Regulatory Particle of the Proteasome from Methanocaldococcus jannaschii.
Mol.Cell, 34, 2009
|
|
4A32
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 3,5-DICHLORO-3'-[(DIETHYLAMINO)METHYL]-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BIPHENYL-4-SULFONAMIDE, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
